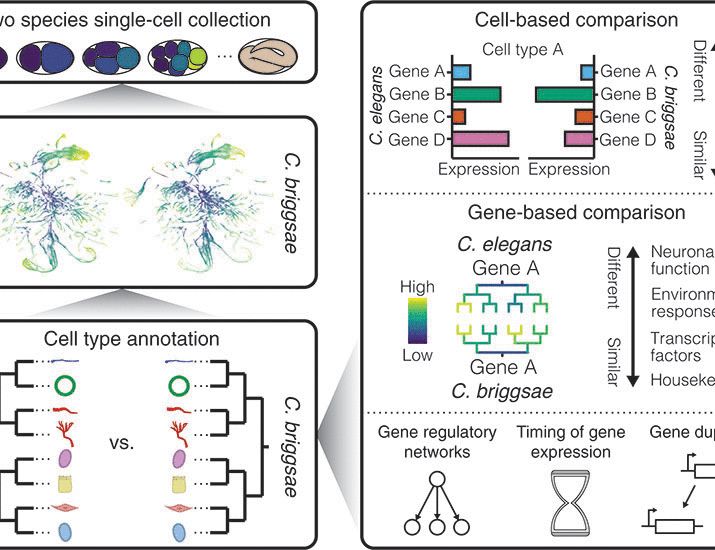
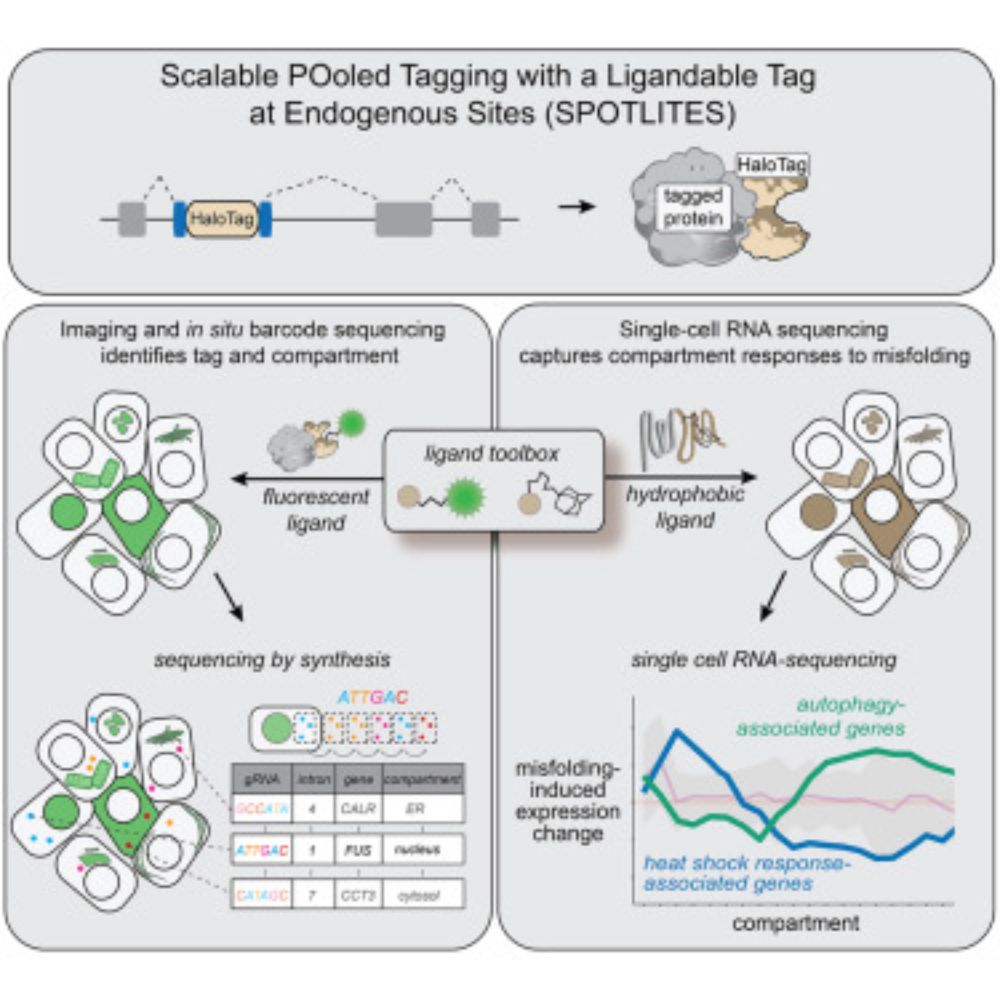
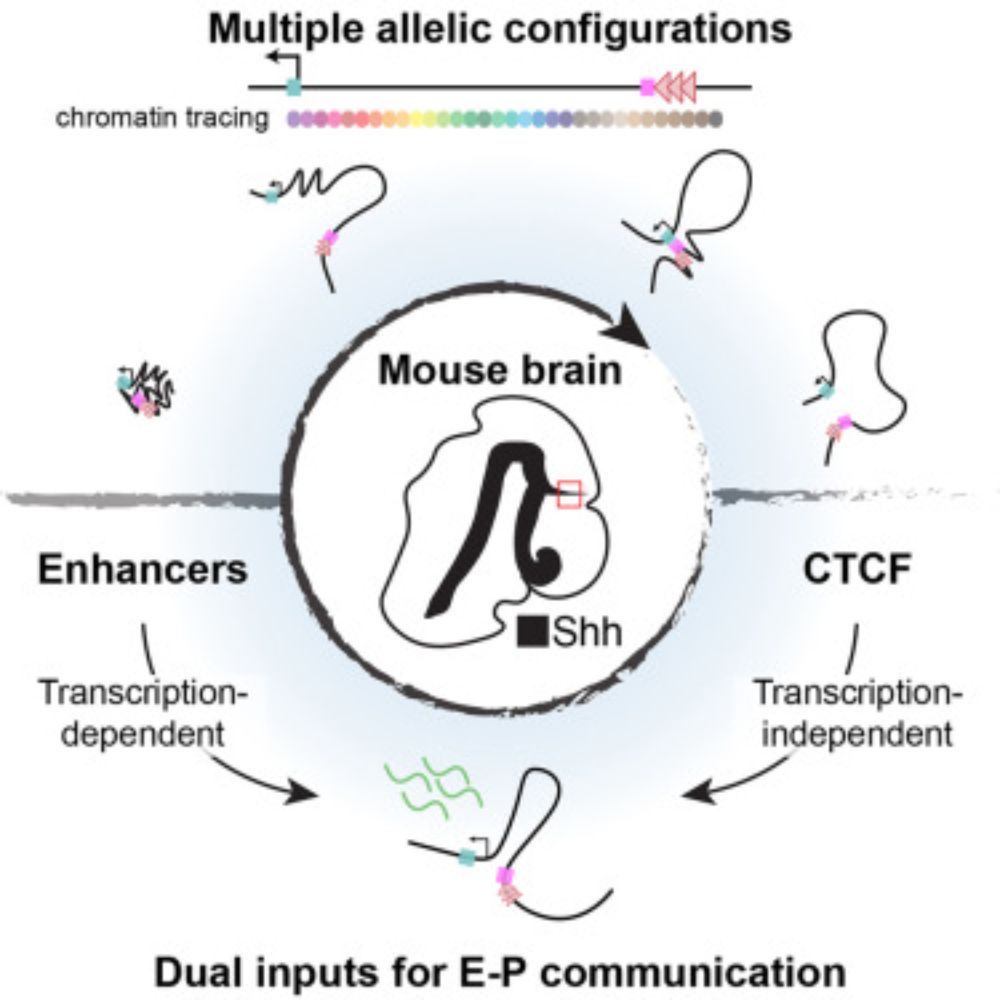
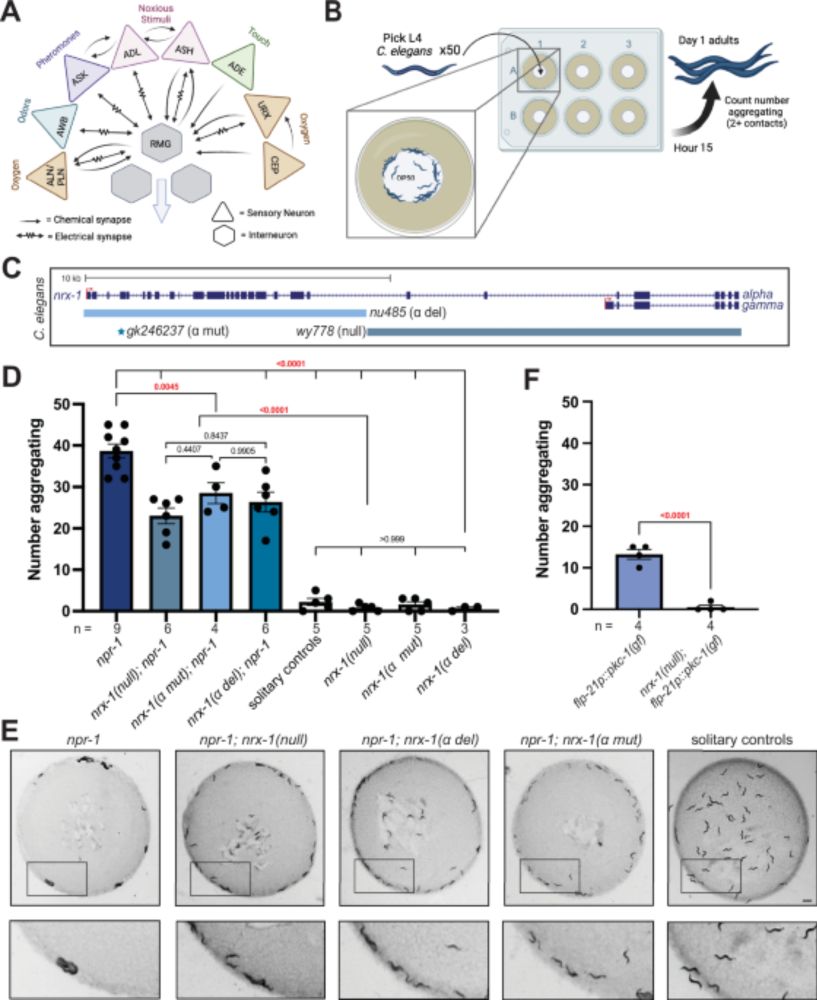
Congratulation @shiraweingarten.bsky.social! Beautiful work!
17.06.2025 18:21 — 👍 2 🔁 1 💬 1 📌 0
Excited to to see our collaboration with @gburslem.bsky.social published today! This is part of our ongoing long term goal of achieving scalable non-disruptive and direct imaging and perturbation of the endogenous proteome
01.05.2025 21:22 — 👍 29 🔁 7 💬 1 📌 0

NimbusImage
Super excited to be launching NimbusImage.com! Cloud-based image analysis to democratize machine learning!
Documentation here!
docs.nimbusimage.com
17.03.2025 14:46 — 👍 91 🔁 34 💬 7 📌 2
We have a postdoc position available for a very cool collaborative project! Synthetic chemistry expertise is a must!
Unfortunately due to the funding source, this position is only available to US citizens or permanent residents.
chemistryjobs.acs.org/job/postdoct...
25.02.2025 21:39 — 👍 4 🔁 3 💬 0 📌 0

Pooled optical screening bootcamp!
05.02.2025 22:06 — 👍 6 🔁 0 💬 0 📌 0
Huge congratulations to former lab postdoc @ysereb.bsky.social for starting his own independent lab! Make sure to follow him for exciting future science and check out his lab if you are looking for opportunities in the NJ/NY area!
23.12.2024 23:05 — 👍 7 🔁 0 💬 0 📌 0
https://www.nature.com/articles/s41556-024-01544-2
Fascinating work from Jongens and @erikaholzbaur.bsky.social labs showing that FMRP granules mark mitochondrial fission sites in axons and dendrites and serve as sites of local translation!
t.co/wXqtte3pqp
15.11.2024 21:13 — 👍 8 🔁 5 💬 0 📌 1
I would guess quite a bit, WPRE is used to stabilize long non terminated transcripts. It is used in lentivirus vectors to compensate for the fact that you cannot use polII termination sequences between the LTRs as this would terminate transcription during lenti production.
10.11.2024 22:04 — 👍 2 🔁 0 💬 0 📌 0
Given that lack of nuclear localization can have a profound impact on editing and gene perturbation efficiency, we suggest further investigation across both cultured and in-vivo postmitotic cell models.
10.11.2024 16:10 — 👍 0 🔁 0 💬 0 📌 0
Addition of a neuronal specific NLS (2X MeCP2) was able to rescue neuronal nuclear localization, expression level and improve activity when sgRNAs were delivered 2 weeks after differentiation
10.11.2024 16:10 — 👍 1 🔁 0 💬 1 📌 0
Other CRISPR enzymes we tested (nCas9 and Zim3) where also found mostly in the cytoplasm of neurons, but were not unstable in neurons
10.11.2024 16:10 — 👍 1 🔁 0 💬 1 📌 0
We found two neuronal specific issues: cytoplasmic mislocalization of CRISPR enzymes after differentiation and the destabilization of the KRAB domain outside the nucleus, which was specific to dCas9-KRAB (using the KOX1 domain).
10.11.2024 16:10 — 👍 0 🔁 0 💬 1 📌 0
This work was spearheaded by Gregory Cajka, brilliant graduate student in the lab, and Matthew Liu, extremely talented undergrad and post-bac research tech.
10.11.2024 16:10 — 👍 0 🔁 0 💬 1 📌 0
Science and fun from the lab of Tony Hyman at the MPI-CBG @mpicbg.bsky.social. Tweets by Hymanlab members.
https://hymanlab.org/
Biophysics, soft matter physics, intrinsically disordered proteins, macromolecular interactions, & phase transitions are my interests. Discovery through rigor and collaboration is my passion. Kindness, generosity, humility, and justice are my hopes.
Our main focus is to offer innovative tools for live cell fluorescence imaging to the scientific community.
https://spirochrome.com/
Official account for the University of Pennsylvania. Advancing discovery and opportunity toward a better future for all. Managed by the Office of University Communications.
Doudna Lab at UC Berkeley, Innovative Genomics Institute founder, CRISPR co-inventor and Nobel laureate innovativegenomics.org/
Exploring how the environment, brain & body interact to shape human health 🔬 | Faculty at Stanford & Core Investigator at @arcinstitute.org
http://www.thaisslab.com
Senior Group Leader IMP / Adjunct Professor Medical University of Vienna. Fascinated by functional genetics (CRISPR, RNAi, degrons) and time-resolved omics towards understanding cancer biology and probing new therapeutic concepts.
Structural biology and pharmacology, with a focus on GPCRs and membrane transporters. 💊 🧪
https://www.gati-lab.com
https://scholar.google.com/citations?user=5t80YUAAAAAJ&hl=en
Engineer/scientist, Nobel Prize in Chemistry 2018
I love evolution, enzymes, protein engineering, AI
Linus Pauling Professor at Caltech
We combine soft matter physics, biophysics and cell biology to uncover physical principles of cellular organization @ Cluster of Excellence Physics of Life, Dresden
Chemist in a CellBio place. Fan of lipids & membranes
Incoming Assistant Professor, Columbia University | @DamonRunyon.org Postdoc, UC Berkeley | PhD Princeton University | Mobile elements, Structural biology and Genome engineering | http://thawanilab.org
Designing peptides/proteins to program biology! 🧬💻🧫 Assistant Professor at Duke | Co-Founder of Gameto and UbiquiTx | MIT SB, SM, PhD
A translational research lab @mskcancercenter.bsky.social focused on the immunobiology and therapeutic potential of genetically engineered #Tcells and #TCRs. Est. 2016 @NYC
Scientist, entrepreneur, ex-biopharm exec, former FM radio jock, lifelong hockey fan, aspiring guitar player and mixologist, #immigrant. CEO, Arrakis Therapeutics and a variety of other things. https://www.michaelgilman.net/disclosures/
The Shechner lab in UW Pharmacology. We study Noncoding RNAs and cellular architecture, and we build “democratized” RNA-focused chemical biology and genomics tools.
He/His/Him. Almost cartainly not D.B. Cooper
ShechnerLab.org
Professor and Director, Dept. of Biophysics & Biophysical Chemistry
Johns Hopkins School of Medicine, structural biologist, lover of chromatin and ubiquitin, fan of the active voice.
Scientist in the Department of Molecular Biology at UT Southwestern Medical Center. We investigate mechanisms of tumorigenesis.
https://labs.utsouthwestern.edu/odonnell-lab
the unculturables: exploratory research & beyond. EMBO-YIP 2003, EAM 2012, adERC 2013, EMBO 2024. https://beja.net.technion.ac.il/
Pediatric genetics physician scientist interested in all things non-coding. https://orcid.org/0000-0002-5409-6103
Views are my own.