
Great culture can save lives. Literally.
Amazing letter in today’s @thetimes.com about Tom Stoppard

Great culture can save lives. Literally.
Amazing letter in today’s @thetimes.com about Tom Stoppard

Excited to see our TIRTL-seq method published in Nature Methods www.nature.com/articles/s41...! Some updates after the revision: (1/4)
25.11.2025 22:38 — 👍 15 🔁 5 💬 1 📌 1Thanks Sander!
25.11.2025 22:05 — 👍 0 🔁 0 💬 0 📌 0
A therapeutic peptide vaccine for fibrolamellar hepatocellular carcinoma @naturemedicine.bsky.social @markyarchoan.bsky.social @hopkinspress.bsky.social @pgtimmune.bsky.social @fredhutch.org @stjude.bsky.social
www.nature.com/articles/s41...

@natmethods.nature.com TIRTL-seq: deep, quantitative and affordable paired TCR repertoire sequencing
www.nature.com/articles/s41...
@pogorely.bsky.social @fredhutch.org @stjude.bsky.social @pgtimmune.bsky.social
We are hiring! We have a postdoc position opened @fredhutch.org to join our team to design next gen cellular therapies for cancer. Heavy focus on T cell signaling and understanding how synthetic receptors instruct T cells to fight cancer.
25.11.2025 01:06 — 👍 10 🔁 9 💬 1 📌 0
FLC work led by @markyarchoan.bsky.social and TIRTL by @pogorely.bsky.social w/ Marina Baretti Allison Kirk,
Jeremy Crawford, Samir Adhikari, @nick-clark-bioinfo.bsky.social, Anastasia Minervina, David Brice, Stefan Schattgen, Zeal Kamdar, & Balaji Sundararaman, Support Fibrolamellar Cancer Found.
If you'd like to try TIRTL, we've got two detailed protocols online, one for the 384-well format (which requires some small volume liquid handlers) and one for a 96-well format which can be done in any lab. protocols.io/view/tirtl-s... and www.protocols.io/view/tirtl-s...
24.11.2025 21:36 — 👍 4 🔁 0 💬 1 📌 0Together, these studies lay groundwork for rational design of epitope-targeted immunotherapies and demonstrate how high-resolution immunomonitoring can guide next-generation T cell therapies, including public motif-driven adoptive cell transfer.
24.11.2025 21:36 — 👍 1 🔁 0 💬 1 📌 0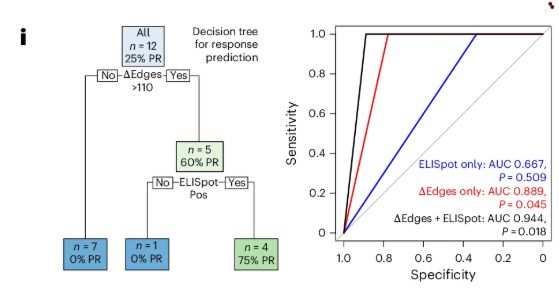
Combining vaccine-induced epitope-specific functional data with TIRTL-Seq repertoire analytics produced a powerful stratification model for clinical response, surpassing standard immune monitoring.
24.11.2025 21:36 — 👍 2 🔁 0 💬 1 📌 0
Applying TIRTL-Seq in our FLC vaccine cohort revealed expansive, polyclonal tumor-reactive T cell responses, many shared across patients with common HLA types. This approach exposed public TCR motifs rarely seen in tumor immunity, and mapped the functional landscape of response.
24.11.2025 21:36 — 👍 2 🔁 0 💬 1 📌 0
A major challenge is that tumor- and pathogen-specific T cells circulate at extremely low frequencies, often below 1 in 100,000 or even 1 in a million, making them essentially invisible to standard single-cell approaches.
24.11.2025 21:36 — 👍 2 🔁 0 💬 1 📌 0
To truly interrogate immune responses at the clonal level, we needed unprecedented resolution. Enter TIRTL-Seq: an affordable, scalable platform generating millions of paired TCRαβ clones per experiment, enabling thorough quantification and tracking of T cell dynamics.
24.11.2025 21:36 — 👍 2 🔁 0 💬 1 📌 0
In a first-of-its-kind trial, we combined the FLC vaccine with immune checkpoint blockade. Among patients with advanced disease, 75% mounted strong fusion-specific T cell responses, with 25% experiencing partial responses and durable progression-free survival.
24.11.2025 21:36 — 👍 2 🔁 0 💬 1 📌 0
FLC is a rare, aggressive liver cancer lacking effective systemic therapies. Our vaccine targets the recurrent DNAJB1::PRKACA fusion, offering a universal neoantigen to drive anti-tumor immunity. www.nature.com/articles/s41...
24.11.2025 21:36 — 👍 5 🔁 1 💬 1 📌 0
Two complementary studies from our group out today: a therapeutic peptide vaccine trial for fibrolamellar carcinoma (FLC) & TIRTL-Seq, our robust method for deep quantitative paired TCR sequencing a🧵w/ @markyarchoan.bsky.social & @pogorely.bsky.social www.nature.com/articles/s41...
24.11.2025 21:36 — 👍 32 🔁 13 💬 4 📌 0Thank you!
07.11.2025 22:24 — 👍 1 🔁 0 💬 0 📌 0Thanks!
07.11.2025 22:24 — 👍 0 🔁 0 💬 0 📌 0Thank you!
07.11.2025 22:24 — 👍 0 🔁 0 💬 0 📌 0Haha yes, thanks!
07.11.2025 22:24 — 👍 1 🔁 0 💬 1 📌 0Thanks Kilian!
07.11.2025 22:23 — 👍 1 🔁 0 💬 0 📌 0
excited that this paper is finally out in @pnas.org :
www.pnas.org/doi/10.1073/...
Led by Gian Marco Visani (effort initiated by Michael Pun), fantastic collaboration with @pgtimmune.bsky.social @asya-minervina.bsky.social and Phil Bradley.

After many rewarding years at @StJudeResearch surrounded by exceptional colleagues & groundbreaking science, I’m delighted to join @fredhutch to push boundaries in viral & cancer immunology. Grateful to St. Jude and looking forward to what’s next at FH! More here: www.fredhutch.org/en/news/cent...
07.11.2025 21:12 — 👍 105 🔁 20 💬 7 📌 0
Viral immunologist @pgtimmune.bsky.social, who recently joined Fred Hutch's Vaccine and Infectious Diseases Division, aims to harness the "incredible potential" of the immune system to advance diagnostics, vaccines and cancer therapies. bit.ly/3WJVpos
07.11.2025 18:08 — 👍 24 🔁 8 💬 0 📌 2Thank you Stacey!! They have all kinds of different birds out here, you need to visit.
07.11.2025 21:10 — 👍 1 🔁 0 💬 0 📌 0This was a collaborative effort between led by Phil Bradley (not on the socials) and @sschattgen, with Kasi Vegesana, @Asya_Minervina, @villanilab, the MGH COVID-19 team, @s_valkiers and many others!
14.07.2025 23:52 — 👍 2 🔁 0 💬 0 📌 0
In short, you can think of MetaCoNGA as our first draft of the human TCR repertoire. Beta code for matching your T cell populations to those from metaCoNGA is available on Github (github.com/phbradley/me...).
14.07.2025 23:52 — 👍 2 🔁 0 💬 1 📌 0Alternatively, maybe you’ve matched TCRs and GEX to a newly curated regulatory unconventional population (previously difficult to match w/TCR sequence). These vary substantially across donors & conditions & may be highly predictive of immune states relevant to health and disease.
14.07.2025 23:52 — 👍 1 🔁 0 💬 1 📌 0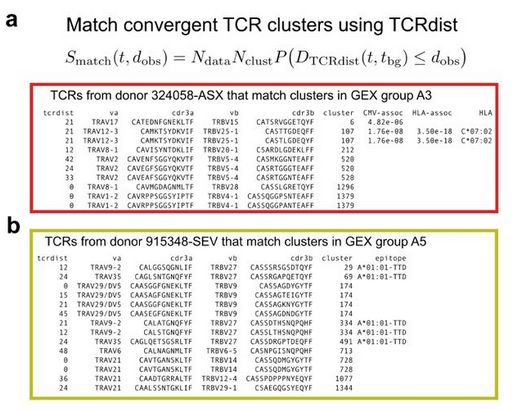
Maybe you’ve identified a novel condition-associated population and you want to see where it falls–is it a conventional epitope specific response? If so, we might be able to tell you the pathogen, the epitope, or the HLA-restriction (or all 3)?
14.07.2025 23:52 — 👍 1 🔁 1 💬 1 📌 0Finally, can we put these two analyses together to make a useful tool for the field? We introduce a mapping tool that allows you to take a new data set and match it to the classifications we’ve defined in MetaCoNGA. This has many uses-
14.07.2025 23:52 — 👍 1 🔁 0 💬 1 📌 0