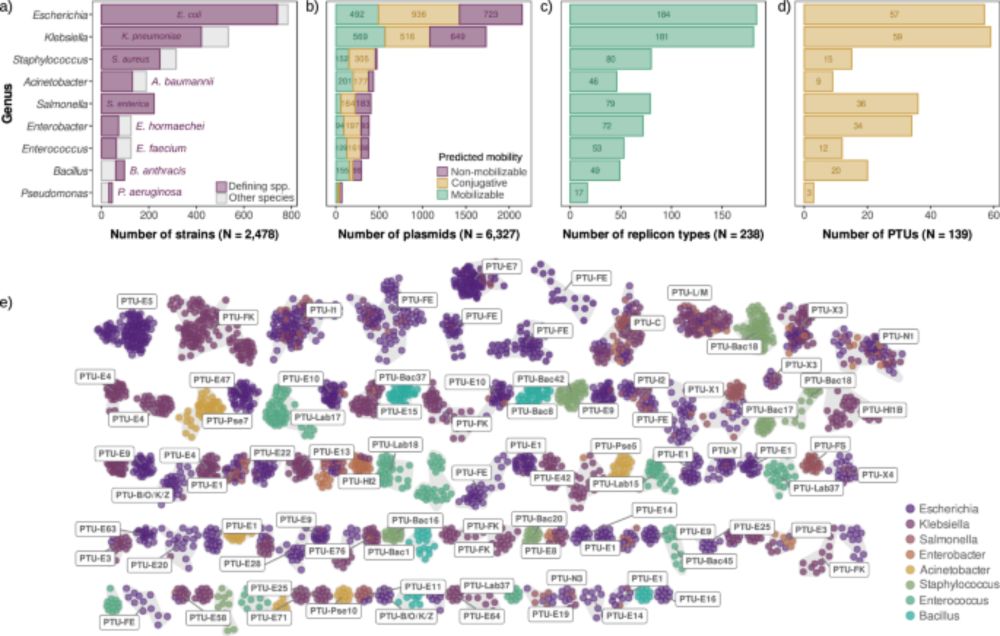
We also found that flight duration matters long-haul flights showed higher ARG transcriptional activity. This underscores the need for targeted surveillance at borders.
04.01.2026 14:11 — 👍 0 🔁 0 💬 0 📌 0
In our new ES&T paper, we analyzed 130 aircraft sewage samples from flights arriving in Saudi Arabia. We identified distinct AMR profiles compared to local sewage, including 51 unique ARGs (like blaNDM, blaVIM).
04.01.2026 14:11 — 👍 0 🔁 0 💬 1 📌 0

Jumbo phage–mediated transduction of genomic islands | PNAS
Bacteria acquire new genes by horizontal gene transfer, typically mediated by mobile
genetic elements (MGEs). While plasmids, bacteriophages, and c...
@prczhaoyansong.bsky.social’s deep dive into the dark matter of compost communities is now out 🎉 Genomic islands hijack jumbo phages—whose capsids enable transfer of large tracts of DNA—shedding new light on the scale & scope of phage-mediated gene flow 😎
www.pnas.org/doi/10.1073/...
28.10.2025 18:36 — 👍 71 🔁 46 💬 1 📌 2

GTDB release 10: a complete and systematic taxonomy for 715 230 bacterial and 17 245 archaeal genomes
Abstract. The Genome Taxonomy Database (GTDB; https://gtdb.ecogenomic.org) provides a phylogenetically consistent and rank normalized genome-based taxonomy
Our @narjournal.bsky.social manuscript is out! It explores the growth of the GTDB (gtdb.ecogenomic.org) since its inception, as well as updates to the website, methodology, policies, and major taxonomic and nomenclatural changes over the past three years.
academic.oup.com/nar/advance-...
22.10.2025 14:20 — 👍 69 🔁 46 💬 0 📌 2

Competitive metagenomic read recruitment explained
A tutorial on the nuts and bolts of competitive read recruitment
I wrote a new tutorial on competitive metagenomic read recruitment (and profiling of the results with #anvio), for those of us nerds who would like to get their hands dirty with data:
anvio.org/tutorials/co...
25.10.2025 10:50 — 👍 39 🔁 9 💬 0 📌 0
I thought I'd try this technical molecular microbiology question here. #MicroSky
Does anyone have experience with cloning 7-10 kbp fragments into a vector for Gram-positives?
We are trying, but the vectors we generally use become quite unstable when we try to clone in a fragment of that size.
11.09.2025 19:52 — 👍 12 🔁 9 💬 8 📌 2

Project - Plasmidlab
PLAS-FIGHTER Exploiting plasmid-bacteria interactions to fight
Antimicrobial Resistance (AMR) could cause 10 million deaths by 2050. At PLAS-FIGHTER, we are working to change this!
🔎 We study how plasmids spread resistance to open up new avenues for future containment and diagnostic strategies.
Find out more 👇
plasmidlab.es/project/
#AntibioticResistance
08.09.2025 08:41 — 👍 19 🔁 8 💬 0 📌 0

Wastewater surveillance tracks spread of antibiotic resistance
A study conducted during COVID-19 travel restrictions sheds light on the relationship between mass gatherings and the spread of antibiotic resistance.
Grateful to KAUST BESE for featuring our Nature Water study on wastewater surveillance of AMR. Huge thanks to Prof. Pei-Ying, our collaborators and BESE community for the support. Excited to keep pushing impactful public-health research forward. #KAUST discovery.kaust.edu.sa/en/article/2...
03.09.2025 12:42 — 👍 1 🔁 0 💬 0 📌 0
New paper in collaboration with the @asantoslopez.bsky.social lab!
www.biorxiv.org/content/10.1...
13.08.2025 06:39 — 👍 25 🔁 15 💬 0 📌 1
This link works: coursesandconferences.wellcomeconnectingscience.org/event/antimi...
22.07.2025 08:50 — 👍 5 🔁 4 💬 0 📌 0

International comparison reveals gender differences in antimicrobial resistance
A recent study led by the University of Turku, Finland, analysed the DNA map of more than 14,000 gut metagenomes in a global dataset and found that there are differences in antibiotic resistance be
International comparison reveals gender differences in antimicrobial resistance - an analysis of more than 14,000 open gut metagenomes indicates that in high-income countries women had more antibiotic resistance than men www.utu.fi/en/news/pres... /w @katammp.bsky.social @mahkamehsalehi.bsky.social
22.06.2025 08:51 — 👍 5 🔁 3 💬 0 📌 0
The deadline for the VIB.AI group leader positions is approaching - send in your CV and short research plan before 14th June to start your BioML research lab in Leuven or Ghent
04.06.2025 07:16 — 👍 10 🔁 10 💬 0 📌 0
PostDoc in ecology and evolution of plasmids in polar waters at HIFMB (f/d/m)
Layout AWI HIPP extern, englisch
We have a new 3-year postdoc position in our group at the @hifmb.de to study plasmids and plasmids systems of the marine environment to survey their utility in microbial responses to environmental change.
Please see the official job ad here, and spread the word:
jobs.awi.de/Vacancies/20...
05.06.2025 08:09 — 👍 79 🔁 85 💬 2 📌 5
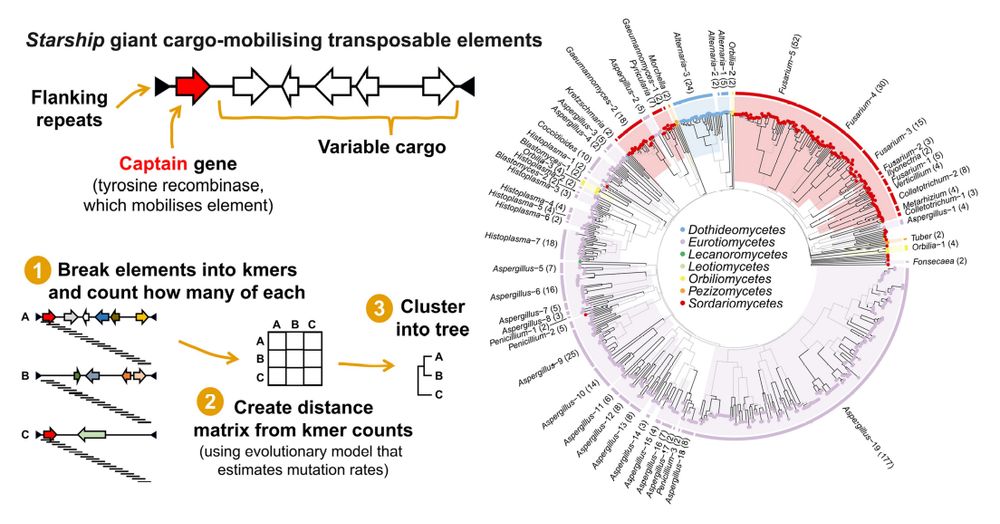
Starship giant transposable elements cluster by host taxonomy using k-mer-based phylogenetics. #TransposableElements #TEs #Kmer #Phylogenetics #Genomics #Bioinformatics #G3 🧬 🖥️
academic.oup.com/g3journal/ad...
02.06.2025 17:06 — 👍 29 🔁 24 💬 0 📌 2

Phased genome assemblies and pangenome graphs of human populations of Japan and Saudi Arabia
The selection of a reference sequence in genome analysis is critical, as it serves as the foundation for all downstream analyses. Recently, the pangenome graph has been proposed as a data model that i...
We made three pangenome graphs 🧬 public, one for the Japanese, one for the Saudi population, and a merged graph (JaSaPaGe). Useful for 🖥️ bioinformatics on either population, or to evaluate how pangenome graphs behave when two different populations are included. jasapage.bio2vec.net/view for PanGene
23.12.2024 09:05 — 👍 7 🔁 2 💬 0 📌 0

Jumbo phage-mediated transduction of genomic islands
Bacteria acquire new genes by horizontal gene transfer (HGT), typically mediated by mobile genetic elements (MGEs). While plasmids, bacteriophages and certain integrative and conjugative elements (ICE...
@prczhaoyansong.bsky.social posted an update of elegant work on genomic islands. Captured via real-time expts, the GIs define a widespread class of MGE (eg VPI-1 de V. cholerae), encode a diverse cargo of genes & exploit jumbo phages for horizontal transfer 😎👇
www.biorxiv.org/content/10.1...
18.05.2025 10:19 — 👍 24 🔁 8 💬 0 📌 1


🎓 Officially passed my PhD defense today. :)
After years of wastewater, metagenomes, and a lot of coffee, it's finally done.
Grateful to my advisor, thesis committee, collaborators, and everyone who supported me along the way.
06.05.2025 14:04 — 👍 1 🔁 0 💬 0 📌 0
Genomes from long-read metagenomic assemblies contain rampant errors, highlighting the pressing need for stricter evaluation methods in long-read assembly algorithms. Read more in our paper with the Eren group. @floriantrigodet.bsky.social @merenbey.bsky.social
28.04.2025 21:29 — 👍 42 🔁 16 💬 0 📌 2
I love both these stories. They are inspiring, and I am also working heavily on metagenomic diagnostics
But this also shows we have made little to zero progress in a decade in using NGS in clinical microbiology and ID, which is depressing
www.bbc.co.uk/news/article...
www.nejm.org/doi/10.1056/...
30.04.2025 12:19 — 👍 8 🔁 3 💬 3 📌 0
Phage-host interaction; Antimicrobial reisistance; Omics
Humboldt Fellow, Helmholtz Center for Environmental Research-UFZ
Associate Professor, Research Center for Eco-Environmental Sciences,Chinese Academy of Sciences
Evolutionary geneticist working with microbes. Absorbed by Major Evolutionary Transitions. Fighting the decline to grumpiness. MPI for Evolutionary Biology, Plön, Germany & ESPCI, Paris, France.
Refugee from Twix. Arsenal fan and Professor of Epidemiology at Harvard T. H. Chan School of Public Health. Opinions my own.
Microbial evolution / University of Manchester
Ecology and evolution in microbes. Assistant Prof. @ LIU Post, native NYer, foodie.
Veterinarian | Resident in Veterinary Microbiology | PhD Candidate | Antimicrobial stewardship obsessed and writer or Resistance is Futile | Passionate about AMR, One Health, infectious disease and acute medicine
Plasmid biology, antimicrobial resistance plasmidlab.es
Editor-in-Chief of a very good microbial journal.
(Illustration by David S. Goodsell, RCSB Protein Data Bank. doi: 10.2210/rcsb_pdb/goodsell-gallery-028)
UKRI-funded research network examining the relationship between AMR, climate change and pollution via a Planetary Health framework. #amr #climatechange
Cares about tiny things in a larger context
Assoc. prof @uniGroningen
Microbiology, ecology, evolution
#AMR, #UTI
microbecoevo.com
Professor and director at the Department of Fundamental Microbiology, University of Lausanne, https://veeninglab.com/. Interested in antibiotic resistance, bacterial cell biology, host-microbe interactions.
Aquatic metabolomics and mass spectrometry. Natural & synthetic compounds in water. At Athens Water (EYDAP SA). Views are our own. www.aquomixlab.com #teammassspec
We are the department of Environmental Engineering and Earth Sciences.
We have 3 programs, Environmental Engineering, Biosystems Engineering, and Earth Sciences. Stay connected for job opportunities, scholarships, news, awards, and much more!
Evolutionary microbiologist interested in AMR & microbiomes 🦠 UKRI Future Leaders Fellow & Group Leader @ Queen’s University Belfast. she/her. Ex-GB athlete 🇬🇧
Prof in Environmental Engineering @EPFL, studying viruses in water and air.
Associate Prof, UC Berkeley. Global health, WASH tech, environmental surveillance, antibiotic resistance, following the pathogens. pickering.berkeley.edu
www.mangrovewater.org
Weekly, peer-reviewed scientific journal on infectious disease epidemiology, prevention & control 😷🩺🧪
#OpenAccess 🔓
#PublicHealth
Publisher: European Centre for Disease Prevention and Control
Impact factor 2024: 7.8
Associate Professor in Computer Science at KAUST. Editor in Chief for the Journal of Biomedical Semantics. Interested in knowledge representation, bioinformatics, neuro-symbolic AI.
Bacterial ecology, evolution and epidemiology | Mathematical and statistical modelling | Genomics | AMR. Assistant professor at the University of Lausanne. https://wp.unil.ch/evolutionaryepidemiology