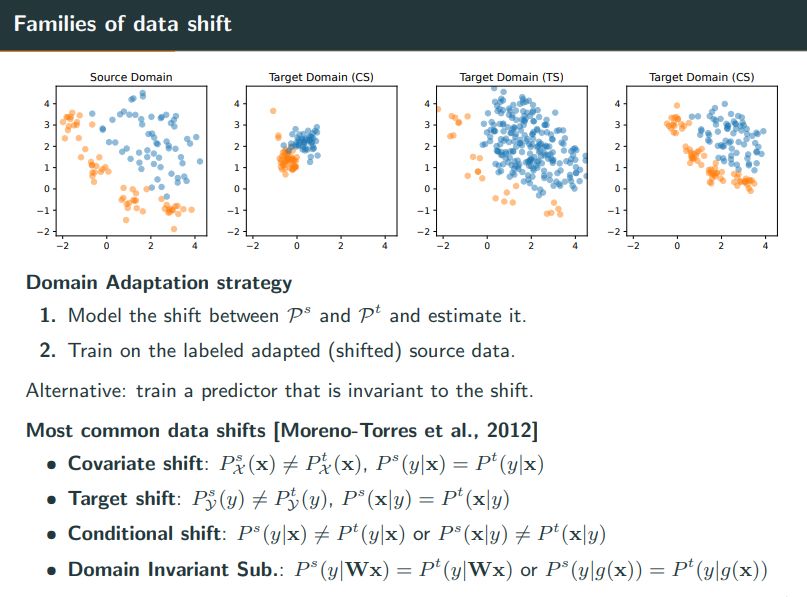
The most important aspect when facing data shift is the type of shift present in the data. I will give below a few examples of shifts and some existing methods to compensate for it.🧵1/6
01.07.2025 09:38 — 👍 30 🔁 16 💬 2 📌 1@rdmichael.bsky.social
PhD student at University of Copenhagen, @DIKU, @KU_BioML group, DDSA fellow.

The most important aspect when facing data shift is the type of shift present in the data. I will give below a few examples of shifts and some existing methods to compensate for it.🧵1/6
01.07.2025 09:38 — 👍 30 🔁 16 💬 2 📌 1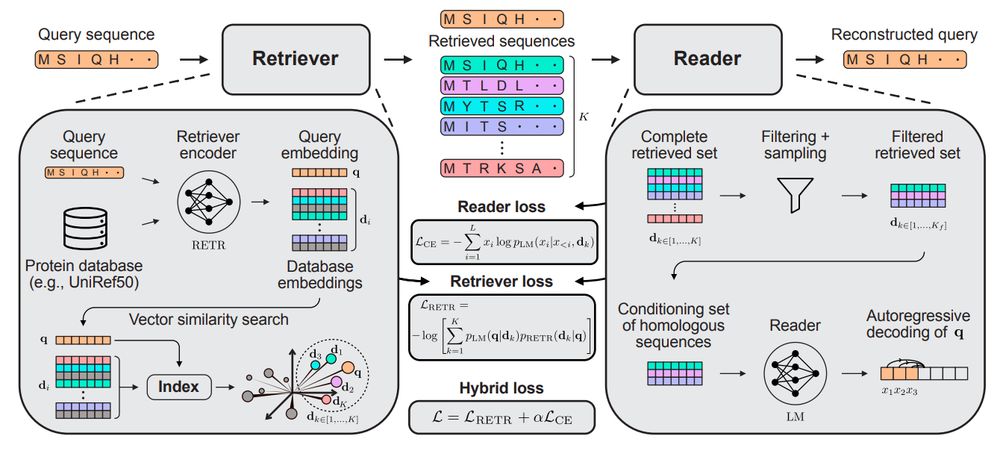
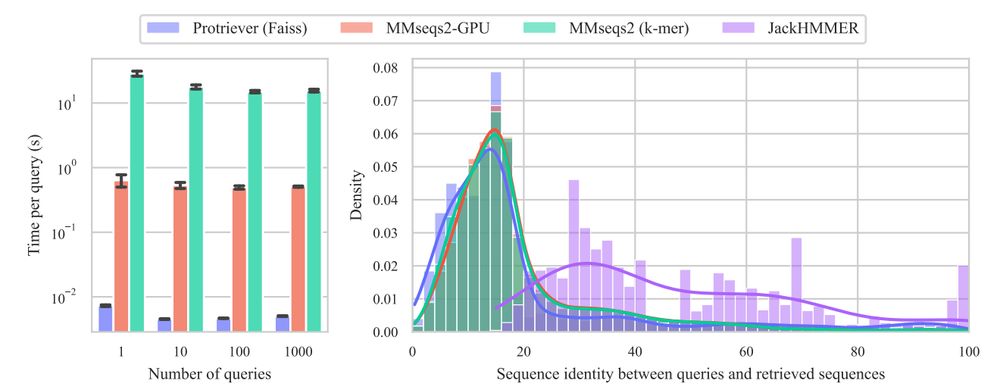
End-to-end differentiable homology search for protein fitness prediction.
@yaringal.bsky.social @deboramarks.bsky.social @pascalnotin.bsky.social
arxiv.org/abs/2506.089...

Spotted Oosterkade in Utrecht
11.03.2025 13:35 — 👍 0 🔁 0 💬 0 📌 0
🚀 It's time for a new JAX ecosystem library!
This time quite a small one: ESM2quinox. A #JAX + Equinox implementation of the ESM2 protein language model.
GitHub: github.com/patrick-kidg...
SciML is obviously my whole jam. My open source has largely focused on the differential equations ...
[1/2]
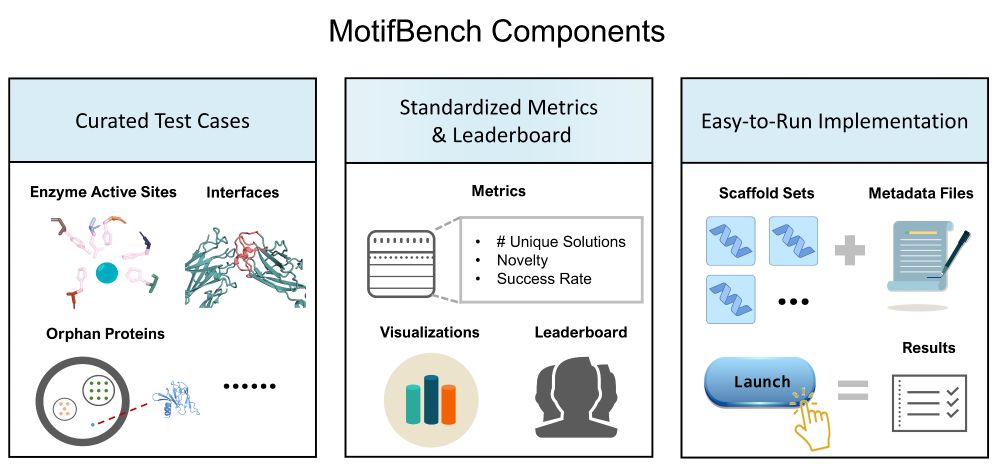
🔥 Benchmark Alert! MotifBench sets a new standard for evaluating protein design methods in motif scaffolding.
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
The BioEmu-1 model and inference code are now public under MIT license!!!
Please go ahead, play with it and let us know if there are issues.
github.com/microsoft/bi...
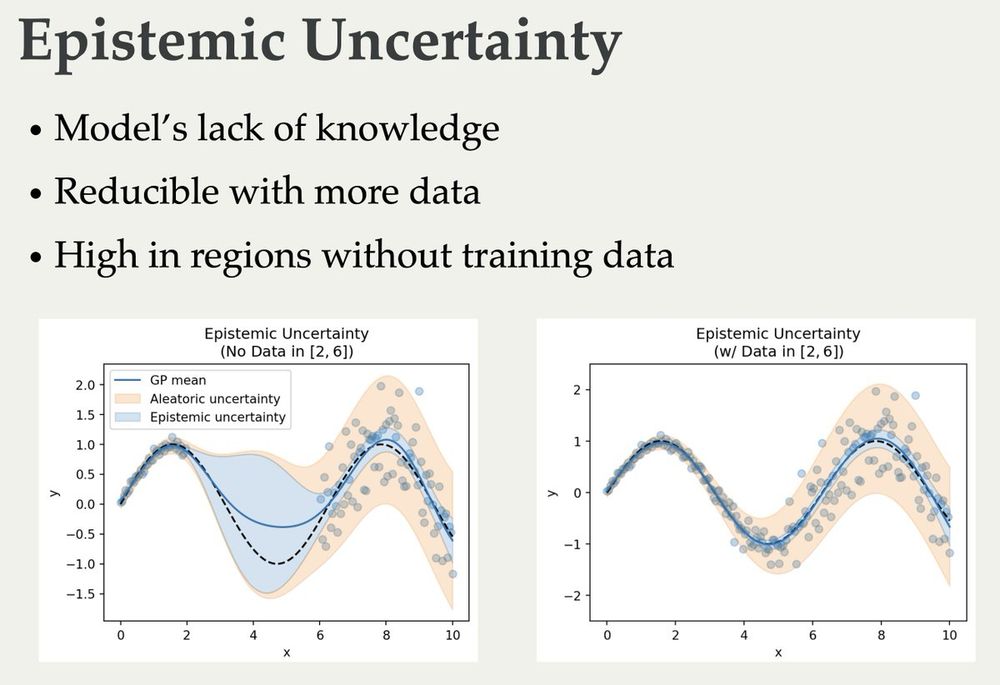
The slides for my lectures on (Bayesian) Active Learning, Information Theory, and Uncertainty are online now 🥳 They cover quite a bit from basic information theory to some recent papers:
blackhc.github.io/balitu/
and I'll try to add proper course notes over time 🤗
I'm very grateful for the work w/ Simon Bartels and @miguelgondu.bsky.social, who did the heavy lifting on the libraries, CI/CD, testing, PR reviews, up-to-date docs, and much more.
Thanks to the generous support and funding by the MLLS center and the #DDSA!
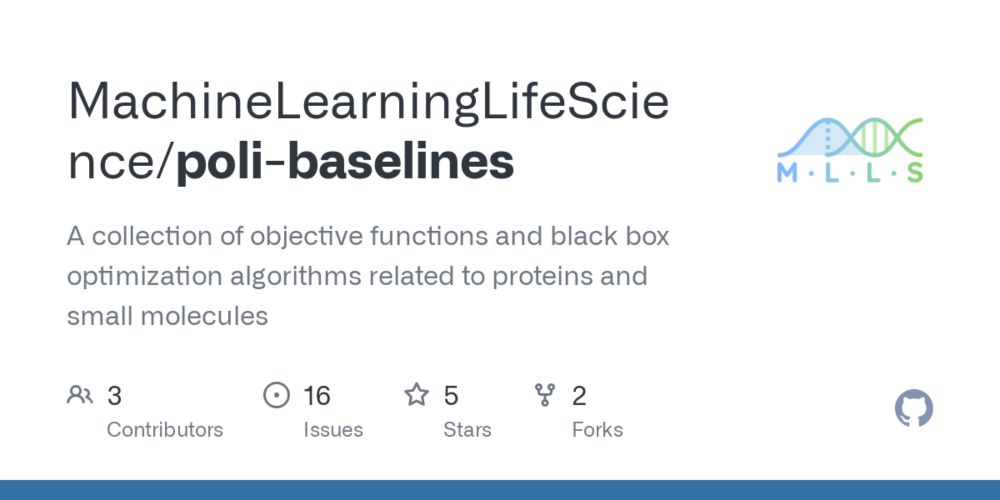
With the tasks in place, we can run and assess different optimizers.
A range of solvers is available in the `poli-baselines` package,
here: github.com/MachineLearn....
The docs on how to get set up and contribute, are here: machinelearninglifescience.github.io/poli-docs/
We developed the `poli` package for easy access to (bio-chemical) black-box functions (cf. TDC, protein stability, ...) and synthetic tasks, with under-the-hood isolation.
Repo: github.com/MachineLearn...
License: MIT (unless a blackbox says otherwise)
A quick install away: `pip install poli-core`
Working on (high-dimensional) Bayesian optimization and care about reproducible, robust comparisons?
Check out our poster presented by @miguelgondu.bsky.social : neurips.cc/virtual/2024... at #NeurIPS2024
Paper: arxiv.org/abs/2406.04739
Site: machinelearninglifescience.github.io/hdbo_benchma...
🧵

Traveling to Copenhagen. Tomorrow, I will give a talk at the DeLTA lab seminar sites.google.com/diku.edu/del...
I will talk about our joint work with Geoffrey Wolfer on the estimation of the average mixing time for Markov chains + consequences for machine learning. Link:
arxiv.org/abs/2402.10506