We (@sobuelow.bsky.social & @kejohansson.bsky.social) tested AF-CALVADOS using the recently described PeptoneBench SAXS benchmark that contains SAXS data for >400 proteins with different amounts of order and disorder. The results look pretty good 😇 so we are sharing here while updating the preprint📝
16.12.2025 15:01 — 👍 21 🔁 8 💬 1 📌 0
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
20.10.2025 11:26 — 👍 93 🔁 37 💬 1 📌 1
Huge thanks to @lindorfflarsen.bsky.social and all the members of SBiNLab at the University of Copenhagen for three fantastic postdoctoral years!
02.09.2025 15:43 — 👍 4 🔁 0 💬 1 📌 0
I’m excited to share that I have started a new position as Senior Scientist, Biomolecular Simulation, at @bindresearch.org in London! We are creating experimental and computational tools and public datasets with the goal of making intrinsically disordered proteins druggable.
02.09.2025 15:43 — 👍 23 🔁 1 💬 2 📌 0
Arriën & Giulio's paper on
A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social
17.07.2025 08:54 — 👍 13 🔁 5 💬 0 📌 0
They mostly leverage things now
25.06.2025 10:12 — 👍 2 🔁 0 💬 1 📌 0
Congratulations! 🙌
23.05.2025 15:33 — 👍 4 🔁 0 💬 0 📌 0
Now published! Big congrats to first author @gginell.bsky.social
We are actively working improving/updating various aspects of FINCHES; don't hesitate to reach out if you run into issues, have questions.
www.science.org/doi/10.1126/...
23.05.2025 11:31 — 👍 121 🔁 46 💬 8 📌 2
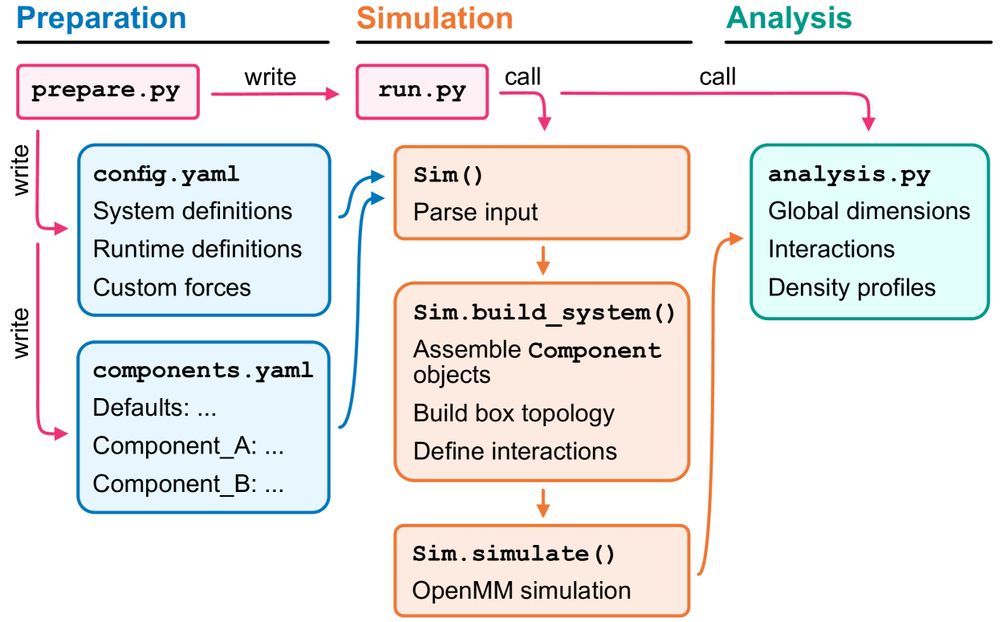
Figure showing the architecture of the CALVADOS package.
Do you like CALVADOS but are not quite sure how to make it?
We’ve got your back!
@sobuelow.bsky.social & @giuliotesei.bsky.social—together with the rest of the team—describe our software for simulations using the CALVADOS models incl. recipes for several applications. 1/5
doi.org/10.48550/arX...
15.04.2025 07:08 — 👍 47 🔁 16 💬 2 📌 2
Supervised training using data generated by multiplexed assays of variant effects is potentially very powerful, but is made difficult by assay- and protein-specific effects
Here @tkschulze.bsky.social devised a strategy to take this into account while training models
www.biorxiv.org/content/10.1...
03.04.2025 03:05 — 👍 27 🔁 6 💬 0 📌 0
Thanks to @lindorfflarsen.bsky.social and all authors for this wonderful project on predicting IDR phase separation from sequence!
Check out the published version (including added exp. data from @tanjamittag.bsky.social) and feel free to try out our webserver.
25.03.2025 18:23 — 👍 23 🔁 4 💬 0 📌 0
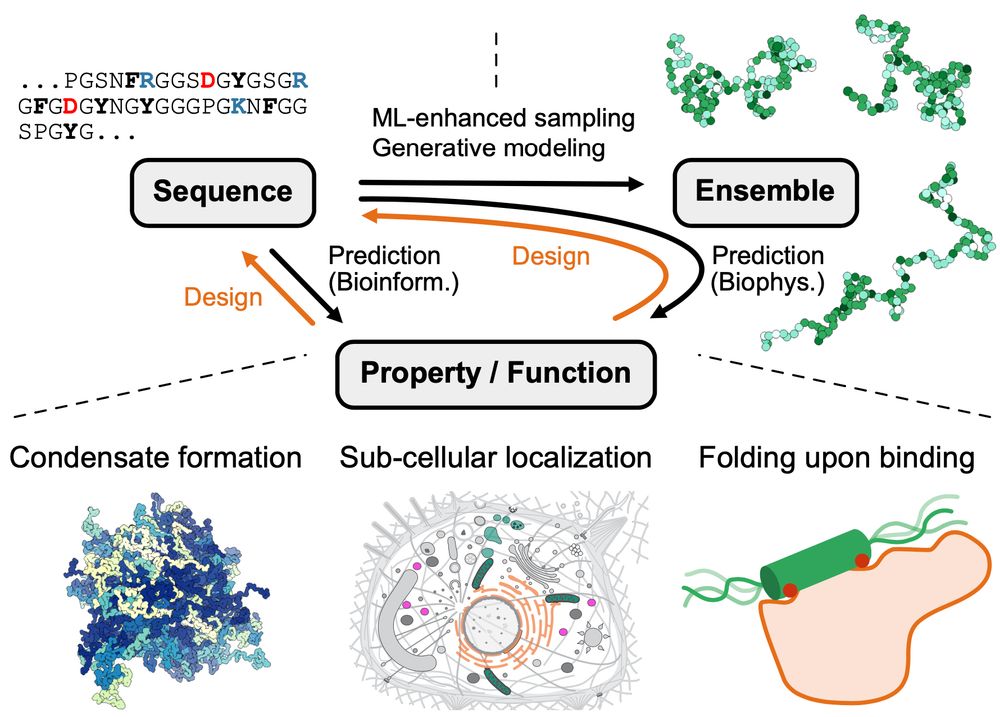
Figure from the paper illustrating sequence–ensemble–function relationships for disordered proteins. ML prediction (black) and design (orange) approaches are highlighted on the connecting arrows. Prediction of properties/functions from sequence (or vice versa, design) can include biophysics approaches via structural ensembles, or bioinformatics approaches via other hetero- geneous sources. The lower panels show examples of properties and functions of IDRs for predictions or design targets. ML, machine learning; IDRs, intrinsically disordered proteins and regions.
Our review on machine learning methods to study sequence–ensemble–function relationships in disordered proteins is now out in COSB
authors.elsevier.com/sd/article/S...
Led by @sobuelow.bsky.social and Giulio Tesei
12.03.2025 21:37 — 👍 91 🔁 27 💬 0 📌 1
CALVADOS 🤝 PEG
Work from @asrauh.bsky.social on a simple model for polyethylene glycol to study the effects of crowding on IDPs
06.03.2025 22:03 — 👍 41 🔁 7 💬 2 📌 2
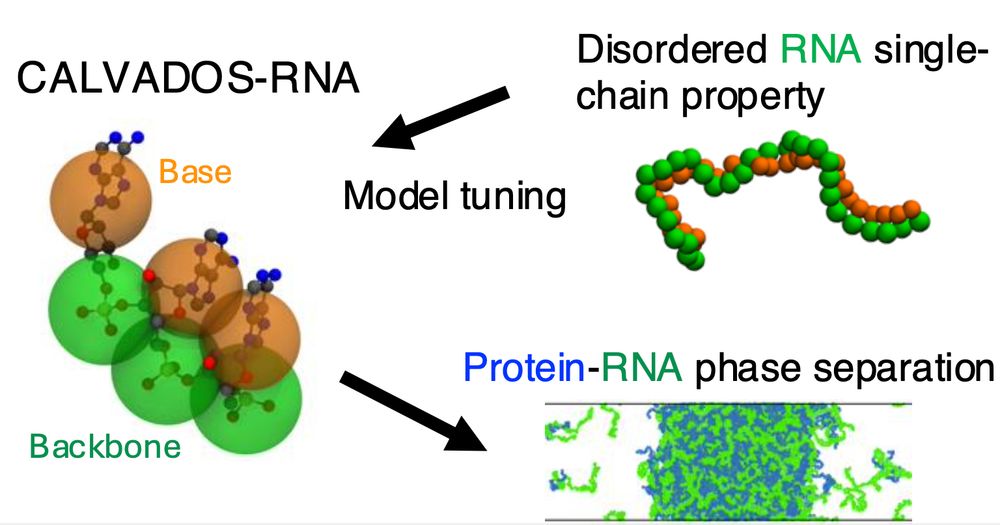
Table of Contents figure showing the CALVADOS-RNA model and a snapshot from a mixed protein-RNA condensate
CALVADOS-RNA is now published
doi.org/10.1021/acs....
This is a simple model for flexible RNA that complements and works with the CALVADOS protein model. Work led by Ikki Yasuda who visited us from Keio University.
Try it yourself using our latest code for CALVADOS
github.com/KULL-Centre/...
26.02.2025 19:11 — 👍 67 🔁 20 💬 1 📌 0
Check out @rasmusnorrild.bsky.social's work with Alex Buell and Joe Rogers developing and using Condensate Partitioning by mRNA-Display to probe phase separation of ~100.000 sequences, and @sobuelow.bsky.social's simulations to support and analyse the experiments
www.biorxiv.org/content/10.1...
21.12.2024 16:32 — 👍 36 🔁 8 💬 1 📌 0
Meet the CALVADOS RNA model
Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪
29.11.2024 08:25 — 👍 89 🔁 21 💬 4 📌 2
BONUS! If IDPs are your jam, check out an ever-expanding starter pack!
go.bsky.app/J23B51L
10.11.2024 19:38 — 👍 30 🔁 19 💬 13 📌 4
New preprint w @tkschulze.bsky.social who analysed cellular abundance (VAMP-seq) data for ~32,000 variants of six proteins 🧪
We find that much of the variation can be explained and predicted by a burial-dependent substitution matrix
Lots more goodies in the paper
doi.org/10.1101/2024...
25.09.2024 16:43 — 👍 8 🔁 2 💬 0 📌 1
“Trust me”
25.09.2024 10:26 — 👍 1 🔁 0 💬 0 📌 0
Happy to share work led by @sobuelow.bsky.social on prediction of phase separation of disordered proteins from sequence
We combined active learning and coarse-grained simulations to develop a machine learning model for quantitative predictions of IDR phase separation 🧬🧶
doi.org/10.1101/2024...
03.06.2024 20:06 — 👍 7 🔁 1 💬 1 📌 0
Updated version of our coarse-grained CALVADOS model 🍎
We show that a Calpha representation of the folded domains can give rise to too compact conformations of multi-domain proteins (MDPs), and that a centre-of-mass representation in the folded domains improves agreement with experiments. 🧬🧶
08.02.2024 09:02 — 👍 23 🔁 10 💬 1 📌 0
Postdoctoral researcher at UC Berkeley | Microalgae, environmental stress, molecular and synthetic biology | Formerly @ Université Paris Sud, Sorbonne Université, AgroParisTech, and ISA Lille.
https://scholar.google.com/citations?user=f5eOF_YAAAAJ
faculty at NYU School of Medicine. We use advanced microscopy techniques to understand gene expression. Opinions my own. timotheelionnet.net
Biophysics for cancer therapies in the Chodera Lab at Memorial Sloan Kettering Cancer Center. Views are my own.
Associate Professor @YaleNeuro • co-Director of Graduate Studies @Yale_INP • @YaleRNA • RNA Neurobiology • Neurodegenerative disease • Immigrant • 🐶 dad • Alum @WhiteheadInst @HopkinsNeuro @PKU1898 🏳️🌈
Assist. Prof. Molecular Modelling
Ghent University
#Protein dynamics #MD simulation
Membrane proteins transporters
Our research concerns biological membranes with particular emphasis on the structure and function of membrane proteins.
#Biophysics #cryoEM #structuralbiology
PhDing at the Sanger Institute, i'm evolving every day
🌼 isabellezane.bio
PhD Student at MPI Biophysics
Computational biophysicist, MD research scientist, & German Wednesday enthusiast. 🇮🇪🇬🇧🇪🇺
Assistant Professor at UCLA
HHMI Hanna Gray Faculty Fellow
PhD: Stanford Biophysics; Postdoc: Broad Institute
Excited about RNA+proteins+IDPs, high-throughput experiments+computation
Scientist. She/her. I like cryoET, fluorescence microscopy, and cell biology.
Developing cryoCLEM methods in the Mahamid lab at EMBL Heidelberg. Formerly studying NPCs in the Beck Lab at MPIBP Frankfurt. MBL Physiology’23 alumn.
Professor in bioinformatics, Stockholm University. Protein structure lover ( interactions predictions, evolution ..). Using machine learning as a part of AI for Sciences for halv my life. In addition to succén, I sometimes rants about sailing or skiing.
🇩🇪 in 🇧🇪 | postdoc at VIB Center for Brain & Disease Research - KU Leuven | chair 2023 VIB postdoc committee | interested in 🧠 neurodegeneration and proteostasis
Computational Biophysicist; Physics Professor & Physics Graduate Director at WFU; Editor-in-Chief, Journal of Biomolecular Structure & Dynamics
Wen Zhu Lab @ Florida State University | Translational Enzymology & Drug Discovery | Student run account | www.wzhulab.com
PhD student at University of Copenhagen, @DIKU, @KU_BioML group, DDSA fellow.
Professor of Theoretical Chemistry @sorbonne-universite.fr & Director @lct-umr7616.bsky.social| Co-Founder & CSO @qubit-pharma.bsky.social | (My Views)
https://piquemalresearch.com | https://tinker-hp.org



