
SAFE-LD: A novel method for the estimation of linkage disequilibrium from summary statistics
Genome-wide association studies (GWAS) have greatly advanced our understanding of the genetic architecture of complex traits. Downstream analyses of GWAS summary statistics require accurate in-sample ...
I am very happy to finally share SAFE-LD, a method convert genotypes to an anonymised privacy-preserving version to be used to precisely compute LD. Led by @gdesanctis.bsky.social Claudia Gianbartolomei @sodbo.bsky.social and Davide bolognini.
📄: shorturl.at/BgXqq
Github: shorturl.at/b49Wa
21.10.2025 09:47 — 👍 23 🔁 10 💬 1 📌 0
 10.02.2025 13:26 — 👍 3 🔁 0 💬 0 📌 0
10.02.2025 13:26 — 👍 3 🔁 0 💬 0 📌 0
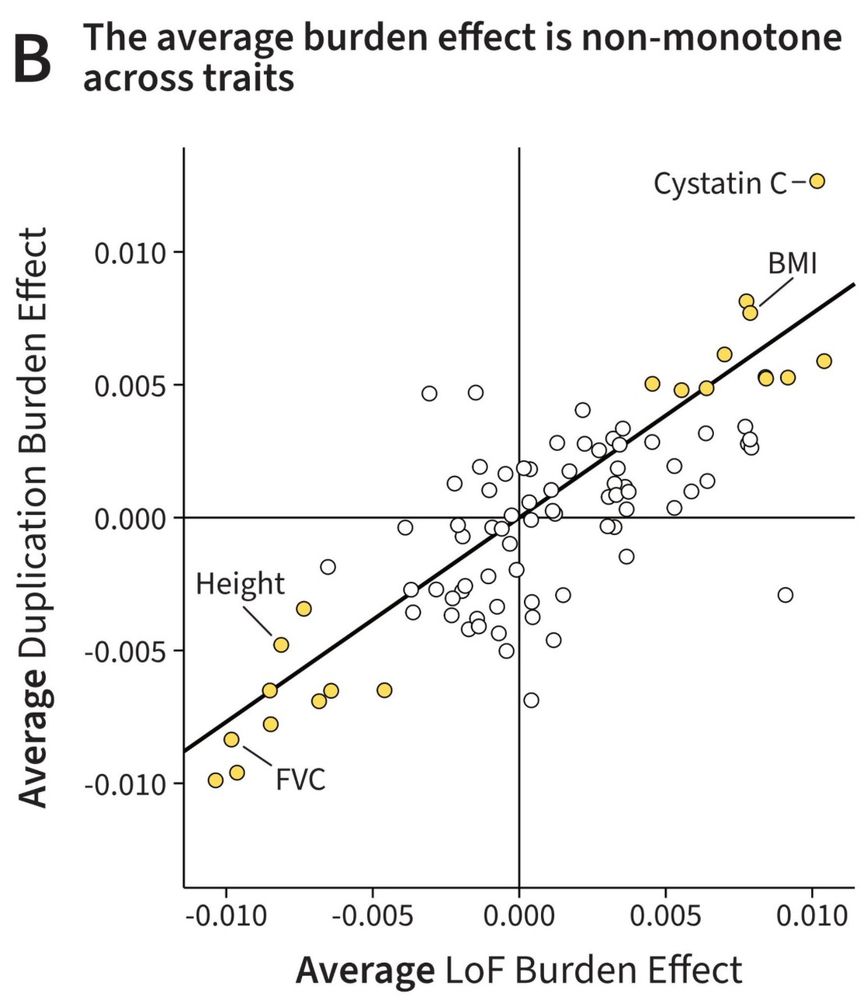
In a new preprint led by @TheNikhilMilind, we explored a fascinating paradox:
For many traits the number of duplications or loss-of-function (LoF) mutations is correlated with phenotype. Curiously, for most traits, the AVERAGE direction of LoFs and Dups is the SAME. Why?
22.11.2024 16:45 — 👍 162 🔁 73 💬 8 📌 4
Beats closest gene!
@jengreitz.bsky.social
@anshulkundaje.bsky.social
And others that I can't find on bluesky. But if you're here let me know so I can follow you!
25.11.2024 00:23 — 👍 51 🔁 16 💬 1 📌 0
Genetics, bioinformatics, comp bio, statistics, data science, open source, open science!
Statistical geneticist. Professor of Human Genetics and Biostatistics at the University of Pittsburgh. Assiduously meticulous.
Assistant Professor at CU Boulder interested in multivariate genomic methods development and their application to understanding shared and unique signal across human disease and risk factors.
Our group develops and applies computational approaches to study molecular variations and their phenotypic consequence. We are part of DKFZ and EMBL.
Website: https://steglelab.org/
Freelance bioinformatician with a focus on human genetics.
Passionate about #GWAS #Metabolomics #Proteomics #Glycomics #Epigenomics, Professor @WeillCornell
Medicine - Qatar, Blog on http://metabolomix.com, http://suhre.fr
🧬 Data Scientist at BioAge Labs
🧬 Visiting Scholar at University of Edinburgh
Longevity drug discovery using causal inference, multi-omics, and large language (of life) models
#Longevity #Aging #Lifespan #Healthspan #MachineLearning #AI
Asst Prof at University of California, Irvine.
Genetics, Genomics, Gene Regulation, Development. Views are my own.
https://www.kvonlab.org/
Associate Prof in health data science @HiLIFE_helsinki @FIMM_UH - MGH/Harvard - Playing with all kind of data - http://dsgelab.org
EMBO Postdoctoral fellow at the Garvan Institute of Medical Research, Sydney, Australia.
Previously EMBL-EBI, Wellcome Sanger Institute and University of Cambridge in Cambridge, UK.
All things single-cell, genetics & genomics.
Genomics, big data, open science, diversity. Director of the Centre for Population Genomics, focused on building a more equitable future for genomic medicine. Opinions my own.
GWAS whisperer.
It's usually the closest gene.
Executive Director, Integrative Biology, Internal Medicine Research Unit. Pfizer R&D. All views my own.
Also cats.
Using statistics and genomics to understand cause and potential treatments for chronic immune mediated diseases.
R, emacs, rake.
Cambridge, UK.
https://chr1swallace.github.io
Computational biologist @HelmholtzMunich, prof @TU_Muenchen & associate PI @sangerinstitute. Dad of 4 and mountain lover. Department news, see @CompHealthMuc
Postdoc fellow @ Stanford & Gladstone Institutes
Core team @scverse-team.bsky.social
Bringing the single-cell genomics in human complex trait genetics
https://emdann.github.io/
Statistical geneticist. Associate Prof at Dana-Farber / Harvard Medical School.
www.gusevlab.org
Complex Trait Genetics | Population Genetics | Evolutionary Genetics
https://scholar.google.nl/citations?user=hsyseKEAAAAJ&hl=en
MBBS, MD, PhD | GWAS storyteller | Scientist at Regeneron | Human genetics & drug discovery in Neuroscience & Psychiatry

 10.02.2025 13:26 — 👍 3 🔁 0 💬 0 📌 0
10.02.2025 13:26 — 👍 3 🔁 0 💬 0 📌 0
