
Bondi senate hearing had a folder full of notes for attacking the senators

Bondi senate hearing had a folder full of notes for attacking the senators

Bondi senate hearing had a folder full of notes for attacking the senators

Bondi senate hearing had a folder full of notes for attacking the senators
Reuters photographer Jonathan Ernst noticed that Bondi flipped open a file during the Senate hearing and he zoomed in.
Inside were her crib notes for attacking the senators.
The GOP now practicing Cliff's Notes Fascism
In other words, under oath Bondi was purely a performance.
07.10.2025 22:47 — 👍 15104 🔁 6483 💬 761 📌 707
Personally, I think a result isn't really worth pursuing unless the underlying protein has been observed in >1000 LC/MS/MS runs & the feature of interest has been observed a few hundred times by multiple groups. But, I am―as everyone knows―more than a bit of an odd ball.
08.10.2025 15:13 — 👍 1 🔁 0 💬 0 📌 0
Michael is involved in a data based effort to do a better job (APPRIS) & I found his enthusiasm for using the preponderance of information from as many sources as possible to be very encouraging.
08.10.2025 14:34 — 👍 0 🔁 0 💬 0 📌 0
For those of you who haven't noticed, I have really enjoyed the posts by @michaeltress.bsky.social in response to my earlier rant involving the use of rules to select special splice variant mRNA (#CanonFolly).
08.10.2025 14:32 — 👍 0 🔁 0 💬 1 📌 0
Maybe HUPO could offer a prize for solving this particular mystery!
07.10.2025 20:31 — 👍 0 🔁 0 💬 0 📌 0
Has anyone ever figured out what is so special/important about the sequence "...SP..." that all chordates have dozens of kinases that only phosphorylate that "S"? #proteomics
07.10.2025 19:43 — 👍 0 🔁 0 💬 1 📌 0

Oracle tumbles after report that it’s lost nearly $100 million from renting out access to Nvidia’s Blackwell chips
Early returns from the AI boom look tiny for Oracle, per The Information....
Bombshell report from The Information is sparking an AI selloff:
Renting out access to Nvidia chips does not appear to be a fantastic money-maker, based on internal docs from Oracle
sherwood.news/markets/orac...
07.10.2025 15:48 — 👍 28 🔁 10 💬 4 📌 1
If any hypothesis you are making requires "all other factors being equal", the sad truth is that they never work out to be that way.
07.10.2025 15:49 — 👍 0 🔁 0 💬 0 📌 0


TSHZ2:p (teashirt zinc finger homeobox 2), apart from a dumb name, is an example of the use of multiple zinc-finger domains on proteins with a single cward homeodomain. In this type of pattern, most of the sequence is structureless, with pS/T acceptors flanking selected zinc fingers. #proteomics
07.10.2025 14:26 — 👍 0 🔁 0 💬 0 📌 0
It could be argued that "Quisling" would be a better parallel, but it is probably a potayto-potahto type situation.
06.10.2025 14:41 — 👍 3 🔁 0 💬 0 📌 0
It is one of those cases where trying to apply rules to select a single translation simply doesn't work in practice.
06.10.2025 14:36 — 👍 0 🔁 0 💬 1 📌 0
I don't know if there is anything you can do at the primary reporting level: whoever uses your data can convert any well-thought out annotation scheme to simple binary. And I would argue that the current literature precedents would strongly encourage that type of transformation.
06.10.2025 12:46 — 👍 1 🔁 0 💬 1 📌 0
Both of these DNA interaction domains act by locating themselves into a major groove in chromosomal DNA. The length of the IDR between them determines how far apart the appropriate grooves can be (in this case adjacent).
06.10.2025 12:18 — 👍 0 🔁 0 💬 0 📌 0


HNF1B:p (hepatocyte nuclear factor 1 homeobox B) has an nward λ repressor-like DNA-binding domain and a nearby cward homeodomain. The human protein only has S/T+phosphoryl acceptors on the N-flank of the 1ˢᵗ domain, but mouse sees fit to add some to the C-flank of the 2ⁿᵈ. #proteomics
06.10.2025 11:59 — 👍 0 🔁 0 💬 1 📌 0
I am always more than a little skeptical when someone characterizes phosphorylation acceptor either observed or not (0/1). Pretty much the definition of a base rate fallacy. #proteomics
05.10.2025 20:12 — 👍 2 🔁 1 💬 1 📌 0
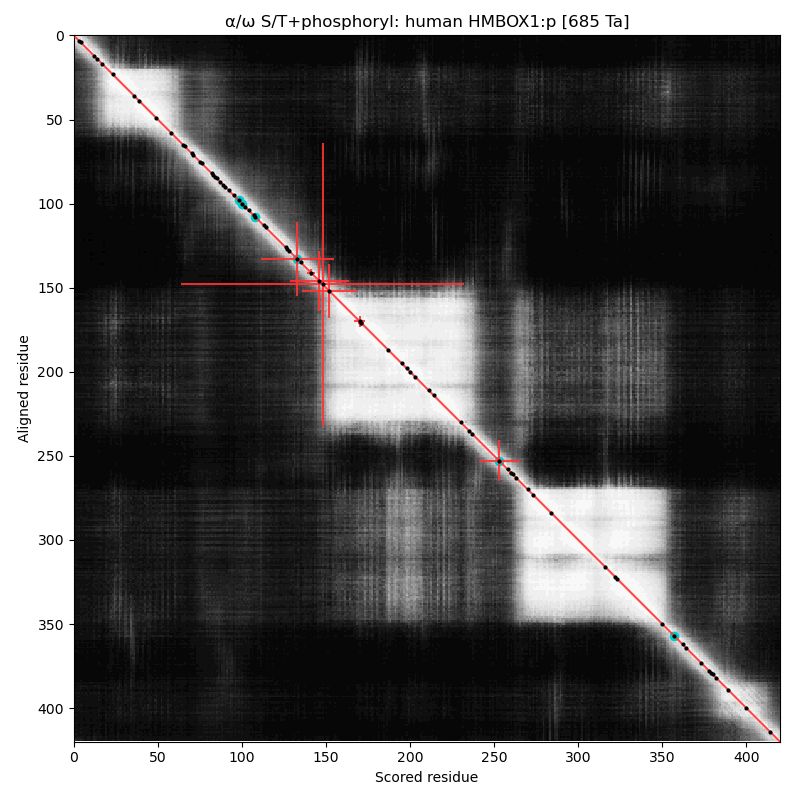

HMBOX1:p (homeobox containing 1) has a cward homeodomain & a central λ repressor-like, DNA-binding domain. Both act together to provide the protein's specific DNA binding, but unlike HOX proteins the occupied S+phosphoryl acceptors sit on the N-flank of the λ repressor-like domain. #proteomics
05.10.2025 12:57 — 👍 0 🔁 0 💬 0 📌 0

3 OCTOBER 2025
Save this map for the books because this is HISTORIC
From 36C in SPAIN to snow in the Balkans
Unbelievable maximum temps <5C at low elevations;
One for all:Kraljevo,SERBIA 215m asl Min 0.4 Max 4.2
with few hours of snow.
Not even Centenarians had ever lived this
03.10.2025 18:54 — 👍 130 🔁 65 💬 2 📌 5
Is it correct to say an imine is "enolizable" if it has alpha protons? I guess an aza-enolate is a thing, so it seems fine to me to say that.
04.10.2025 00:26 — 👍 13 🔁 3 💬 5 📌 0

Like all other HOX family member proteins, the homeodomain of HOXB6:p is located near the C-terminal & has two short helices oriented at right angles to a longer helix that interacts with specific DNA sequences.
04.10.2025 15:14 — 👍 0 🔁 0 💬 0 📌 0

Typhoon Matmo is still on course to make landfall in southern China and northern Vietnam, running along the coast of the Gulf of Tonkin.
04.10.2025 13:32 — 👍 0 🔁 0 💬 0 📌 0

Another day of #aurora powered by an active sun and the Russell-McPherron effect.
04.10.2025 13:28 — 👍 0 🔁 0 💬 0 📌 0


HOXB6:p (homeobox protein family B, member 6) has occupied S/T phosphorylation acceptors flanking its cward homeodomain in both directions. It is an ortholog of fruit fly Antennapedia (Antp), but is not involved in antenna formation in mammals. #proteomics
04.10.2025 12:47 — 👍 0 🔁 0 💬 1 📌 0
APPRIS agrees with UniProtKB on this one, but MANE is right - the inserted exon has no proteomics or RNAseq support. We have changed the APPRIS principal.
03.10.2025 21:16 — 👍 1 🔁 1 💬 0 📌 0
Most of the older generations of proteomics folks tend to believe that exons are a bit of a myth & that UNIPROT canons are unquestionably correct.
03.10.2025 20:40 — 👍 0 🔁 0 💬 1 📌 0
The little rant I went on (canonfolly) was meant to interest at least some of the younger proteomics folk in mRNA assemblies by showing that it leads to observable consequences in MS-based proteomics data. I was not expecting to interest someone from the mRNA world in proteomics data.
03.10.2025 20:39 — 👍 0 🔁 0 💬 2 📌 0
Yeah, it is good to see that there is some cleaning up going on.
03.10.2025 20:02 — 👍 1 🔁 1 💬 0 📌 0
Old person forgetting everything they ever knew about proteomics at speeds in excess of 270 Hz...
Your least favourite person's least favourite award-winning journalist✨
Follow for short-form videos on issues that matter! 💅
SUBSCRIBE: rachelgilmore.substack.com/
https://linktr.ee/rachel_gilmore
FT columnist. Also have podcast. Author of three and a half books. Four kids. Two cats. One wife. Numerous friends. Fulham fan.
Scientist studying molecules using mass spectrometry. Also coding sometimes.
ORCID: 0000-0002-5865-8994
Professor of Proteomics. Currently at Newcastle University UK. he/him
Here in personal capacity.
Proteomics, humor, bbq, gravel riding, hiking, violin, gardening. Durham, NC
Focused (now) on analyzing whole 🩸 in sepsis. Pulmonary, Allergy and Critical Care Medicine, and Proteomics and Metabolomics Core Facility, Duke University
Scientist, still hanging around the UC Davis Genome Center. I guess I currently have 11 protons, because people keep telling me i'm Sodium Fine ;)
#massspectrometry, #proteomics, and #bioinformatics | hiking and dogs | co-founder & CTO @talusbio | opinions my own
A nerd at large. Writer. Musician.
Sometimes funny. 🏳️🌈
Join my sci-fi book club:
http://patreon.com/ellecordova
nerd humor • space & sci fi • books
This is the official account of the Technical University of Munich – Technische Universität München (TUM).
Posts from the web communications team at TUM Corporate Communications Center
Website: http://tum.de/en
Legal notice: http://tum.de/legal-notice
Senior Lecturer in Political Science @POLSISEngage, researching leadership, followership, populist movements, voter attitudes and political behaviour.
https://scholar.google.com.au/citations?user=GwUV0YAAAAAJ&hl=en&oi=ao
Official account for APTN News, our stories told our way.
Immunology & RNA Biology at the University of Vermont. Troubleshooting biology is my happy place. http://biolab.dev // It's all fun and games and "Reg, transporting really is the safest way to travel" until somebody gets bitten by quasi-energy microbes.
A tech news podcast to help you sort through the wreckage.
Hosted by @parismarx.com and @bcmerchant.bsky.social. Music by @com.y-a-c-h-t.com. Production by @ericwickham.ca.
Professor in Biomolecular Mass Spectrometry and Proteomics, Utrecht University, the Netherlands
Spread too thinly. Toronto. Originally from UK. Cancer biology researcher at Sinai Health. Also Prez of the Terry Fox Research Institute.
Assistant professor in genomic and big data, Animal Science department, Laval University, Canada. Opinions are my own. il/lui he/him
https://paquetlab.fsaa.ulaval.ca/
mass spectrometrist and instrumentation rascal, representing myself and not my institution
ElonJet | Tracking Elon Musk’s Jet by @jacks.grndcntrl.net
Also follow @spacexjets.grndcntrl.net




















