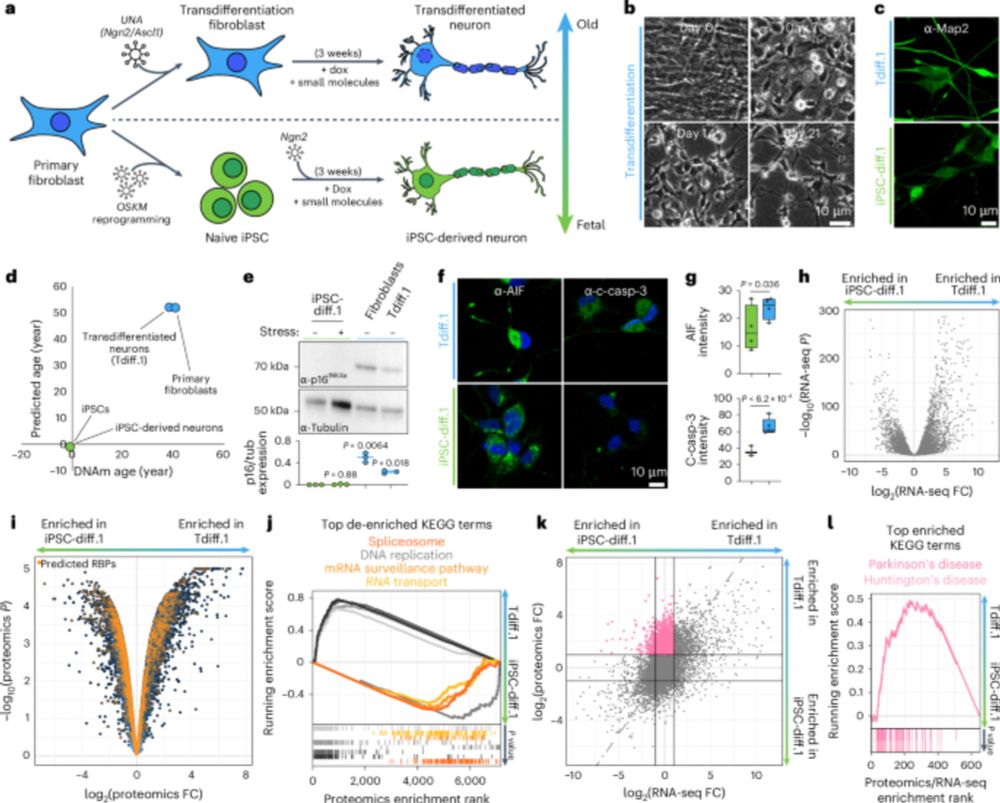
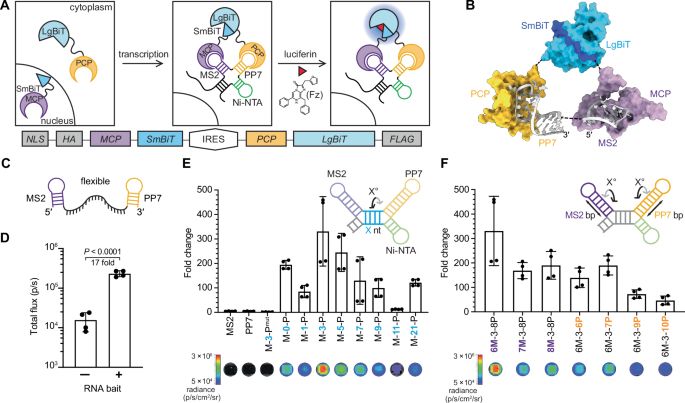
Excited to share our work with @markrcookson.bsky.social and Payel Sen labs. We combine #SingleCell and #Nanopore sequencing to profile #RNA isoforms from young to geriatric mouse brains. We uncover #aging changes in transcription, processing, and splicing that also impact coding potential.
#RNAsky
09.06.2025 16:37 — 👍 6 🔁 0 💬 0 📌 0
🚨 We’re looking for a curious mind to join our lab at the NIH!
Passionate about #RNA biology, #MachineLearning, and the #genomics of #aging?
Interested in a postdoc or a PhD research visit at NIH? Let’s connect.
📩 DM or email for details!
#Bioinformatics #AI #ComputationalBiology
04.06.2025 17:30 — 👍 5 🔁 6 💬 0 📌 0

Single-molecule multimodal timing of in vivo mRNA synthesis
mRNA synthesis requires extensive pre-mRNA maturation, the organisation of which remains unclear. Here, we directly sequence pre-mRNA without metabolic labelling or amplification to resolve transcript...
The timeline of co-transcriptional #mRNA processing and #modification may have to be revised: cleavage, termination, and #m6A deposition may occur earlier than most #splicing! Potential textbook changing study in bioRxiv by edueyras.bsky.social & Rippei Hayashi labs!
www.biorxiv.org/content/10.1...
29.04.2025 20:31 — 👍 55 🔁 21 💬 1 📌 1
Yeah almost all pop up when I allow the word mRNA. Also cleaning up some myself. Hopefully it doesn't get too much out of hand.
10.04.2025 22:54 — 👍 4 🔁 0 💬 0 📌 0
It is based on regular expressions, not tags, so anything (with some filters :) ) with the word RNA in some context will make it to the feed. If signal to noise ratio gets lower, then stricter filters may be needed.
10.04.2025 16:32 — 👍 2 🔁 0 💬 1 📌 0
I've built an #RNA feed for personal use to capture RNA biology posts across the entire network. Some arbitrary filters used to hopefully increase signal to noise ratio but otherwise not moderated. Feel free to follow if you find useful.
10.04.2025 16:09 — 👍 17 🔁 5 💬 2 📌 0
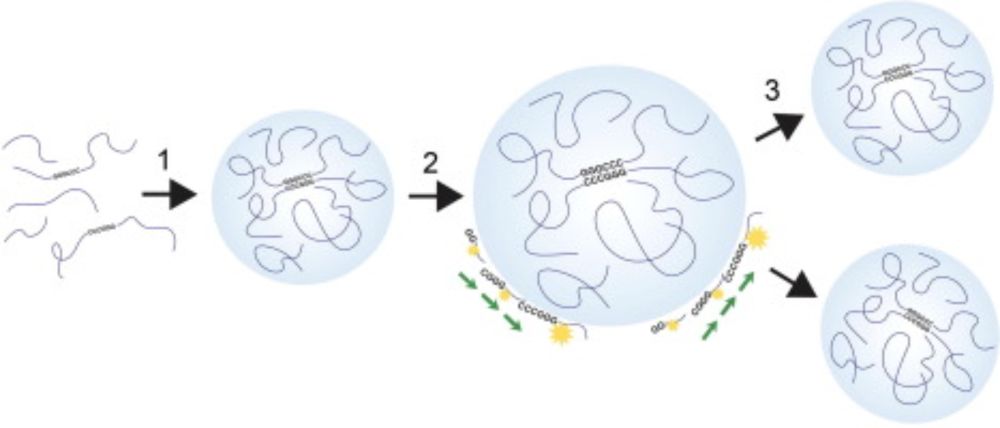
An RNA condensate model for the origin of life
The RNA World hypothesis predicts that self-replicating RNAs evolved before DNA genomes and coded proteins. Despite widespread support for the RNA Wor…
An interesting perspective and framework: #RNA condensates-not individual RNAs-may have acted as replicative units in early life. RNAs in condensate recruit similar RNAs, then the condensate divides once it reaches a critical size.
#RNAsky
10.04.2025 00:28 — 👍 20 🔁 10 💬 0 📌 1
Latest work from our next door neighbors! #RNAsky #RNAbiology
12.02.2025 22:49 — 👍 14 🔁 4 💬 0 📌 0

2022-2025 X Twitter mention data
Here's Twitter's descent since 2022
06.02.2025 09:15 — 👍 324 🔁 92 💬 7 📌 32
Thanks Eugene!
22.12.2024 01:12 — 👍 2 🔁 0 💬 0 📌 0

Our paper out in eLife. Full-length direct RNA #nanopore sequencing uncovers 5' #RNA shortening upon cellular #stress that depends on ✅ stress-granules and #XRN1 but ❌ not poly(A) or cap removal elifesciences.org/articles/96284
🤔 useful to further reduce translational load during #ISR?
#RNAbiology
21.12.2024 01:36 — 👍 84 🔁 23 💬 4 📌 2
SERVICE UPDATE
We normally don't ask you to "share like and subscribe" but please distribute this.
You are now able to delete the link after it embeds on your post so you can get those precious extra characters, and we will catch your post mentioning research.
#AcademicSky #Medsky #HigherEd
11.12.2024 15:00 — 👍 278 🔁 147 💬 9 📌 11
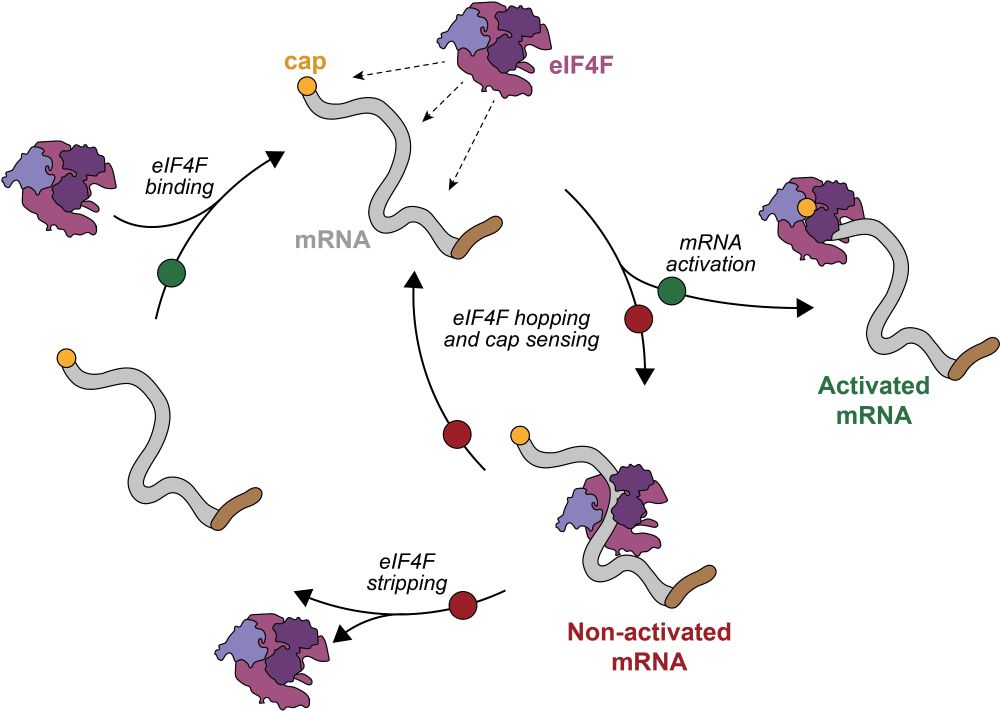
Our latest work is out in Nature today! Using smFRET, we directly visualized recruitment of the eIF4F complex to the 5' cap of eukaryotic mRNAs and formation of an activated mRNA. Our findings reveal new and surprising roles for each eIF4F component. 1/3 www.nature.com/articles/s41...
11.12.2024 21:35 — 👍 172 🔁 55 💬 11 📌 4
Thanks. 👀 seems I'm the most permanent staff. Coming from snakemake, maybe time to dive into nextflow as well 😅
12.12.2024 01:07 — 👍 0 🔁 0 💬 0 📌 0
Yes I do use snakemake regularly but that's only because I found it a bit easier for my workflow several years back. Mostly HPC.
12.12.2024 01:01 — 👍 1 🔁 0 💬 0 📌 0
Not looking to start a debate on #nextflow vs. #snakemake, but curious about their ease of adoption in a typical lab setting with ~4-year PhD/postdoc turnover. How do you feel newcomers adapt to each? Would you recommend one for easier onboarding? Feedback appreciated!
#genomics #bioinformatics
11.12.2024 05:36 — 👍 6 🔁 0 💬 4 📌 0

How can physics define universal laws for computation, from cells and chips to brains? In this @pnas.org Perspective, led by @sfiscience.bsky.social David Wolpert and Jan Korbel, they suggest how Stochastic Thermodynamics can provide the proper framework pnas.org/doi/epub/10....
07.12.2024 12:42 — 👍 49 🔁 15 💬 0 📌 0
🎄🧪 Racing to submit your manuscript before the holidays? Check out our ShinyApp for editorial insights on publishers & journals—submission-to-acceptance times and more! Based on data from: doi.org/10.1162/qss_...
Check the app here 👉 pagoba.shinyapps.io/strain_explo...
#AcademicSky
#Science
07.12.2024 10:05 — 👍 23 🔁 5 💬 1 📌 1
#RNAsky #RNAbiology
04.12.2024 12:52 — 👍 5 🔁 2 💬 0 📌 0
Genomics, Computational Biology, Professor of Stat and Data Science @ Penn
computational biologist and data scientist
jeneaiadams.com
blackwomencompbio.org
linkedin.com/in/jeneaadams
Views my own.
David Bartel's lab @WhiteheadInst @MIT @HHMI | microRNAs, mRNAs, and other RNAs
Biology PhD Candidate at MIT studying lysosomal RNA degradation. UC Berkeley Alum
Senior Scientist that investigates the molecular mechanisms that regulate skeletal muscle mass and function in health and in various disease states. Protein synthesis, ubiquitin biology.
https://scholar.google.com.au/citations?user=ICAcUqUAAAAJ&hl=en
PhD | Biomedical Scientist 👩🔬🔬🧬
Cancer Research | RNA and Molecular Biology
Where Cell-Cell adhesion meets RNAi - and other intriguing encounters. Molecular and Cellular Biology Lab at the Medical University of South Carolina in Charleston.
https://medicine.musc.edu/departments/regenerative-medicine/research/kourtidis-lab
The leading provider of open source workflow orchestration software for data pipelines, cloud infrastructure & collaboration. From the creators of @nextflow.io
Since 1911, we have supported the molecular bioscience community, promoting the sharing of knowledge and enhancing career development.
Discover more about the Society: https://linktr.ee/biochemicalsociety
MSc. Neuroscience &MSc.Physiotherapy| Just a researcher, microglia lover |I hope to become a PhD student in neuroscience | Neuroscience, Anatomy & Exercise Physiology
American immigrant in Sweden working as a statistician at the National Board of Health and Welfare. This is my English language account. #VoteBlue #BLM
The Gregory lab @DukeU studies disease mechanism using scRNAseq & spatial molecular technologies. Focus on brain tumors, Alzheimer's and MS 🧠 Home to the Molecular Genomics Core (MGC) - https://dmpi.duke.edu/cores/molecular-genomics-core-mgc
Scientist at UChicago, Department of Biochemistry and Molecular Biology.
Assistant professor at the Interdisciplinary Nanoscience Center, Aarhus University in Denmark and genetically related to the origin of life.
I study functional nucleic acids focusing on their role in starting evolution.
Nucleic Acids Research (NAR), from Oxford University Press, publishes the results of leading-edge research into physical, chemical, biochemical and biological aspects of nucleic acids and proteins involved in nucleic acid metabolism and/or interactions
Research group in the Biochemistry department @cambiochem.bsky.social at the University of Cambridge exploring the RNA Universe! 🧬✨ https://www.ericmiskalab.org/
Managed by lab members
Organisers of @cambridgerna.bsky.social
Genomics, Bioinformatics.
github.com/tobiasrausch
Postdoc @altmeyerlab. Interested in Checkpoint maintenance, DNA replication, DNA repair & Imaging.