
more links: universityofcalifornia.quorum.us/campaign/sta... universityofcalifornia.quorum.us/campaign/sci...
15.08.2025 19:35 — 👍 0 🔁 0 💬 0 📌 0@konopkalab.bsky.social
Official Konopka Lab Account! We're a group of spirited scientists exploring the genetic landscape of the evolved human brain. @Los Angeles, CA, konopkalab.org

more links: universityofcalifornia.quorum.us/campaign/sta... universityofcalifornia.quorum.us/campaign/sci...
15.08.2025 19:35 — 👍 0 🔁 0 💬 0 📌 0
The future of science is at risk, we need your help to stand up for science: The federal government’s $1B demand from @UCLA puts critical research, innovation, and public health at risk. Join us in standing up for UC’s mission and future. #StandUpForUC: www.universityofcalifornia.edu/get-involved...
15.08.2025 19:33 — 👍 2 🔁 0 💬 1 📌 0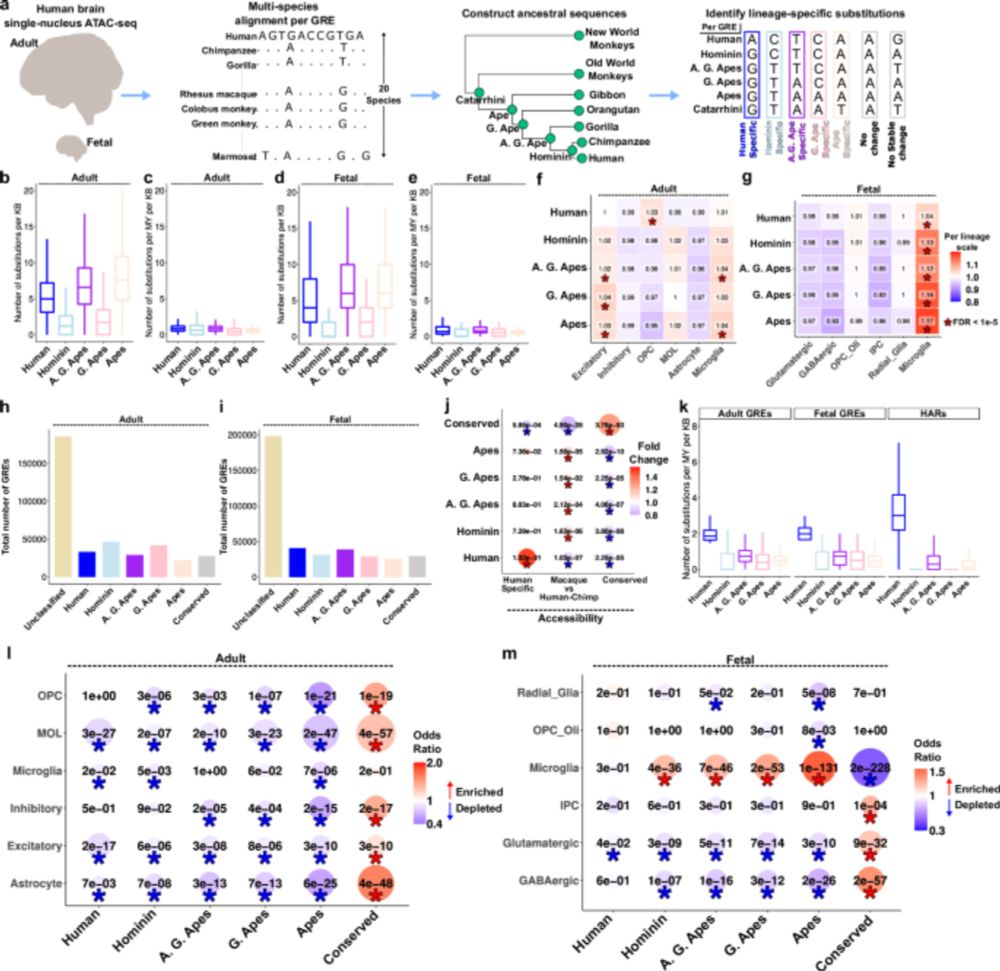
Revealing ancestral evolutionary patterns of the human brain at cell-type resolution, through comparative genomic analysis of open-chromatin regions across apes, old world monkeys & new world monkeys. New integrative paper from @konopkalab.bsky.social in @natcomms.nature.com. 🧠🧬🧪
04.07.2025 17:45 — 👍 29 🔁 9 💬 0 📌 0
Human CLOCK enhances neocortical function @natneuro.nature.com @konopkalab.bsky.social @circadianclocks.bsky.social
www.nature.com/articles/s41...


We celebrated Haley graduating from the Neuroscience Ph.D. program at UT Southwestern. Huge congrats, Dr. Moore! 🎉🎓
16.07.2025 19:31 — 👍 0 🔁 0 💬 0 📌 0
Big news—we’ve moved to the David Geffen School of Medicine at UCLA! Even bigger: Gena is now Chair of the Department of Neurobiology. Congrats, Dr. Konopka! 💙🧠 #UCLA #Neurobiology #NewBeginnings
16.07.2025 18:55 — 👍 6 🔁 0 💬 2 📌 0Thanks for the love! 🌟 Unfortunately, we don't have data from the ventral striatum at the moment, but who knows what the future holds? Always exciting to think about the possibilities! 😄
23.04.2025 15:08 — 👍 1 🔁 0 💬 0 📌 08/
🧪 Combining snRNA-seq + smFISH + cross-species comparisons + cross region comparisons = powerful insights into mammalian striatum evolution.
🧵Thanks for reading!
7/
Takeaways:
🧠 Our findings underscore the importance of including a diverse array of species and brain regions when uncovering novel cell types and examining cellular abundance variations which may lead to species-specific behaviors.
6/
We’re sharing the first snRNA-seq datasets from:
🐵 Chimpanzee CN (n=4) + Pu (n=6)
🦇 Bat (Phyllostomus discolor) CN + Pu (n=4 each)
Available to the neuroscience community! 🌍
5/
Uncovered species-specific differences in interneuron cell type abundance:
PDGFD PTHLH PVALB– interneurons = primarily found in primates 🐒
PDGFD PTHLH PVALB+ interneurons = primarily found in non-primates 🦇🐭🦦
Evolutionary divergence in PVALB expression within striatal interneurons!
4/
🦇 Having analyzed bat (Phyllostomus discolor) CN and Pu snRNA-seq separately we observed:
🔍 Significant neuron-to-glia ratio differences between CN and Pu.
📌2 novel bat interneuron types in Pu which may account for species-specific behavior:
🔹 LMO3 expressing
🔹 FOXP2 and TSHZ2 expressing
3/
🤯 Across ~94 million years of evolution, we found shared molecular markers for eccentric spiny neurons (eSPNs) in both primates 🐒 and non-primates 🐭🦦🦇.
These conserved markers suggest that eSPNs are a core striatal feature, as previously shown in mouse, not a recent innovation.
2/
Eccentric spiny neurons (eSPNs):
Found in lower proportions among SPNs in non-primates 🐭🦦🦇.
In primates, higher eSPN prevalence may support advanced cognitive functions and complex behaviors.
✅ Again, validated with smFISH. 🔬
1/
In primates, the CN shows lower neuron-to-glia ratios vs. non-primates.
This matches known allometric scaling of neuron density in larger brains
🧠 Bigger brains = ↓ neuron density but relatively stable glial density.
(See: Hirter et al. 2021, Herculano-Houzel 2014)
✅ Confirmed with smFISH. 🔬
We generated new snRNA-seq data from primates 🐒, and bats 🦇 as well as used other data from marmosets and non-primates 🐭🦦 from the caudate nucleus (CN) and putamen (Pu) (caudoputamen for mouse), uncovering fascinating differences in cell types and ratios. Let’s dive in! 👇
22.04.2025 17:32 — 👍 0 🔁 0 💬 1 📌 0
We are super excited to share our latest pre-print in collaboration with @sonjavernes.bsky.social lab! www.biorxiv.org/content/10.1... : Evolution of the cellular landscape in mammalian striatum 🧠🧬🧵
22.04.2025 17:32 — 👍 12 🔁 4 💬 2 📌 0Hi Cedric! Thank you for highlighting our work! Stay posted for more cool studies from our lab!
22.04.2025 14:26 — 👍 1 🔁 0 💬 0 📌 0
Very nice comparative study by the labs of Gena Konopka & Sonja Vernes, on the evolution of the cellular landscape in mammalian striatum 🧪🧠🧬
www.biorxiv.org/content/10.1...