Philadelphia Freedom. 🇺🇸 #NoKings
18.10.2025 19:04 — 👍 21033 🔁 5261 💬 388 📌 226

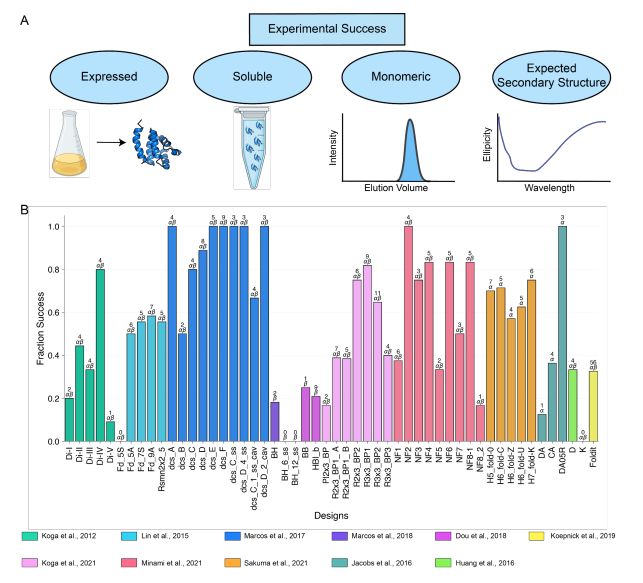
Structures from AlphaFold3 - while often impressively good - tend to fail representing the dynamic ensembles accurately. And often parts of the structure are not correct.
Adding experimental data, directly in AlphaFold's diffusion step, provides physically realistic protein ensembles. This image shows two cases where AlphaFold3-only structures were largely improved by guiding with experimental data.
📢 New preprint:
Experiment-guided AlphaFold3 resolves accurate protein ensembles.
doi.org/10.1101/2025...
AlphaFold3 is incredible, but has crucial limitations: it typically collapses to a single conformation, ignoring the inherent dynamics of proteins. And it can be wrong. Here's a solution. 🧵👇
18.10.2025 18:59 — 👍 52 🔁 22 💬 1 📌 0

BIG ANNOUNCEMENT📣: I haven’t been this excited to be part of something new in 15 years… Thrilled to reveal the passion project I’ve been working on for the past year and a half!🙀🥳 (thread 👇)
15.10.2025 12:22 — 👍 482 🔁 185 💬 55 📌 59
YouTube video by ProteinDesignStudio
Screen binders using ipSAE
pip install ipsae
from www.linkedin.com/in/ullah-sam...
www.youtube.com/watch?v=A5ph...
PyPI pypi.org/project/ipsae/
His github fork github.com/ullahsamee/I...
My github github.com/DunbrackLab/...
Paper www.biorxiv.org/content/10.1...
For designed protein binders www.biorxiv.org/content/10.1...
16.09.2025 19:21 — 👍 20 🔁 6 💬 1 📌 1
Brought to you by Austin Cheng (@auhcheng.bsky.social) — meet the newest member of our team: Quetzal!
Named after Quetzalcoatl, the Aztec god of creation, Quetzal is a simple yet scalable model for building 3D molecules atom by atom.
📜 arxiv.org/abs/2505.13791
[1/4]
23.05.2025 16:13 — 👍 12 🔁 6 💬 1 📌 2
Exciting to see our protein binder design pipeline BindCraft published in its final form in @Nature ! This has been an amazing collaborative effort with Lennart, Christian, @sokrypton.org, Bruno and many other amazing lab members and collaborators.
www.nature.com/articles/s41...
27.08.2025 16:14 — 👍 304 🔁 109 💬 14 📌 11
New preprint: www.biorxiv.org/content/10.1...
We used molecular dynamics simulations and solid-state NMR (@fmp-berlin.de) to look at calcium binding to the pore domain of the calcium-gated K+ channel MthK. @wojciechkopec.bsky.social @compbiophys.bsky.social
19.08.2025 07:42 — 👍 17 🔁 6 💬 1 📌 2

ACS announces 2026 national award winners
The winners are being acknowledged for their outstanding achievements in chemistry across various fields in the discipline
ACS announces 2026 national award winners
The winners are being acknowledged for their outstanding achievements in chemistry across various fields in the discipline. cen.acs.org/people/award...
18.08.2025 20:25 — 👍 14 🔁 4 💬 0 📌 3

CACHE 7 is launched with support from the @gatesfoundation.bsky.social and unpublished data from Damian Young at @bcmhouston.bsky.social, Tim Willson @thesgc.bsky.social and Neelagandan Kamaria InSTEM. Design selective PGK2 inhibitors. We'll test them experimentally.
bit.ly/4lnVYOs
12.08.2025 19:07 — 👍 10 🔁 6 💬 0 📌 0

We are delighted to announce that our perspective article, “Steering towards safe self-driving laboratories (SDLs),” has been accepted for publication in Nature Reviews Chemistry.
Link: www.nature.com/articles/s41...
18.08.2025 17:46 — 👍 15 🔁 7 💬 1 📌 1
Thrilled our PANCS-binder tech is finally out in print!
www.nature.com/articles/s41...
This technology, 11-years in the making, has transformed how we do research in my lab, and was led by an amazing postdoc, Matt Styles, who is currently entering the academic job market - so look out for him! 1/n
06.08.2025 17:54 — 👍 40 🔁 9 💬 4 📌 0

Scaling down protein language modeling with MSA Pairformer
Recent efforts in protein language modeling have focused on scaling single-sequence models and their training data, requiring vast compute resources that limit accessibility. Although models that use ...
Excited to share work with
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
05.08.2025 06:29 — 👍 95 🔁 43 💬 1 📌 1
ChimeraX daily builds can predict binding affinity of small molecules using Boltz 2 on your Mac, Windows or Linux computer. www.rbvi.ucsf.edu/chimerax/dat...
24.07.2025 02:44 — 👍 84 🔁 18 💬 1 📌 4
when they close it all up, I do believe this will go down as the funniest video on the internet
24.07.2025 19:51 — 👍 19772 🔁 4396 💬 672 📌 612
Super excited 3 days at the CECAM workshop co-organized with @drgregbowman.bsky.social @bettinagkeller.bsky.social! Protein dynamics is definitely shaping up to be the next big topic in AI for Science! Huge thanks to @cecamevents.bsky.social, @uwdsi.bsky.social, TCI, Chem, Emma and the DSI staff!
19.07.2025 17:04 — 👍 6 🔁 3 💬 1 📌 0
This was a great meeting! Kudos to @xuhuihuangchem.bsky.social and UW for being such great hosts. Now to talk with the dean to get lake and brat stand at Temple...
18.07.2025 15:53 — 👍 3 🔁 0 💬 1 📌 0

This week @uwdsi.bsky.social is hosting the @cecamevents.bsky.social Flagship Workshop on
Biomolecular Dynamics in the Age of Machine Learning, organized by DSI affiliate @xuhuihuangchem.bsky.social @drgregbowman.bsky.social & Bettina Keller @uwmadisonchem.bsky.social
www.cecam.org/workshop-det...
15.07.2025 12:44 — 👍 8 🔁 4 💬 0 📌 1
Apply - Interfolio
{{$ctrl.$state.data.pageTitle}} - Apply - Interfolio
I am on a glide path to retirement. So my colleagues are looking for a new physical/analytical chemist. I have had a terrific 40 year run at Bryn Mawr…and will miss the students a ton. #chemsky #womeninSTEM apply.interfolio.com/170426
16.07.2025 17:02 — 👍 45 🔁 14 💬 3 📌 2
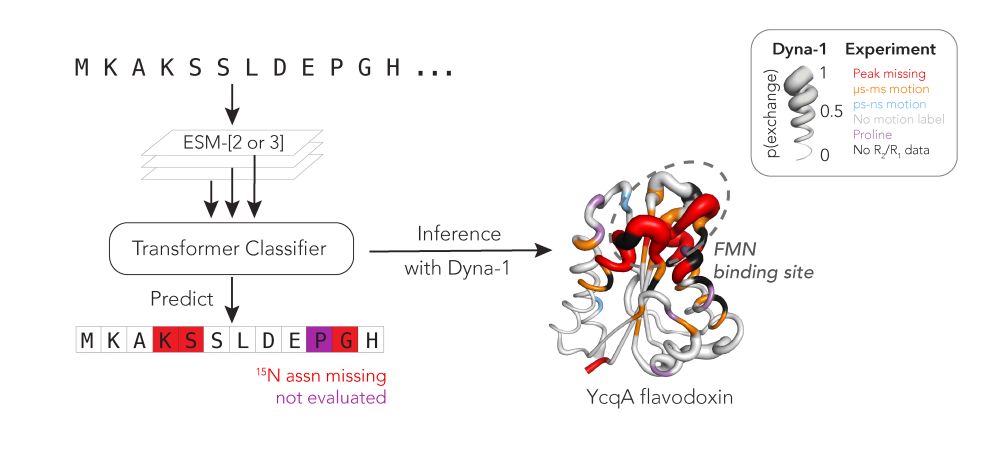
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics?
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
20.03.2025 15:02 — 👍 103 🔁 38 💬 6 📌 5
BioEmu now published in @science.org !!
What is BioEmu? Check out this video:
youtu.be/LStKhWcL0VE?...
10.07.2025 18:57 — 👍 89 🔁 28 💬 6 📌 4
https://vaikuntanathan-group.uchicago.edu
TV & standup & improv & podcasts, hardly any movies
paulftompkins.com/live
Computational Biochemistry | CovinoLab
Assistant Prof @UBChemistry | Computational modeling of RNA structure and folding, intrinsically disordered proteins and phase separation | BioSimLabUB.github.io
About TEs, IFN, senescence with a zest of ML
🇫🇷 Parisian gravitating in the Pacific Northwest 🇺🇲
-> Seattlelite🗼🛸👾
Computational Biochem/Biophys/Pharm Scientist
PDRA at LAMMASU-UFCSPA
Curating Magnetic Resonance news since 2011. use #NMRchat (New Magnetic Resonance Chat) or tag 🧲 for everything MR, NMR, EPR, ESR and MRI. No endorsements
Upcoming Magnetic Resonance Events in 2025/26 (google doc online, being populated) t.ly/4laEY
Biophysicist, skating fan, husband, dad. Santa Fe, New Mexico, USA.
Theory and Computation team leader in Max Planck Institute of Immunobiology and Epigenetics
Postdoc in the Cole Group at Newcastle University interested in molecular mechanics force field development and free energy calculations.
PhD student at MPINAT. Microtubules and coarse grained simulations.
I'm a pharmaceutical chemist working at the interface of cheminformatics, molecular modeling, and machine learning. All views are my own.
https://ligandintheloop.blogspot.com/
Phase separation and disordered proteins!
source: https://arxiv.org/rss/stat.ML
maintainer: @tmaehara.bsky.social
Group Leader
The Genome Function Laboratory
The Francis Crick Institute, London
Computational Biology PhD Candidate - Genome Function lab @crick.ac.uk | 💻 🧬 Functional genomics | Genome-wide
Associate Professor @ Big Data Institute, University of Oxford
2024 Lister Institute Fellow
genomics | rare disease | gene regulation | genetic therapies
https://rarediseasegenomics.org/
(field) hockey player | cyclist | hiker
Research in AI for Protein Design @Harvard | Prev. CS PhD @UniofOxford, Maths & Physics @Polytechnique