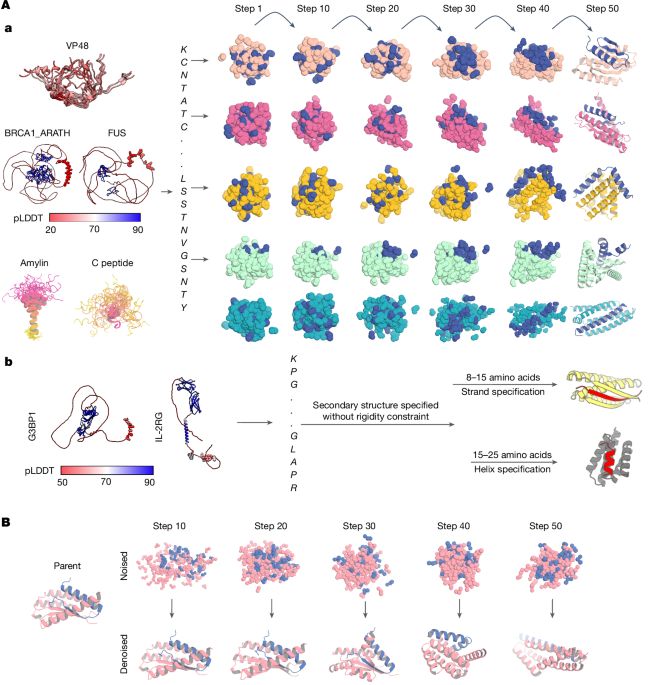
Our review on Integrative modelling of biomolecular dynamics 💃 is now live at COSB
We discuss approaches to integrate computational methods with time-dependent & time-resolved experiments to study protein dynamics
With @dariagusew.bsky.social & Carl G. Henning Hansen
doi.org/10.1016/j.sb...
09.12.2025 22:13 — 👍 56 🔁 20 💬 1 📌 0
Independent Research Group Leaders (f/m/d) SCALE / FIAS
The Frankfurt Institute for Advanced Studies (FIAS) and the newly funded Cluster of Excellence SCALE are seeking several Independent Research Group Leaders to drive theoretical and computational research in molecular and cellular life science.
fias.institute/de/das-fias/...
05.12.2025 17:30 — 👍 11 🔁 10 💬 0 📌 0

Across (Conformational) Space and (Relaxation) Time: Using Coarse-Grain Simulations to Probe the Intra- and Interdomain Dynamics of the Tau Protein
The biological importance of intrinsically disordered proteins (IDPs) has been established for over two decades, yet these systems remain difficult to characterize, as they are better described by conformational ensembles instead of a single reference structure for their folded counterparts. Tau is a prominent member of the IDP family, which sees its cellular function regulated by multiple phosphorylation sites and whose hyperphosphorylation is involved in neurodegenerative diseases such as Alzheimer’s. We used coarse-grain MD simulations with the CALVADOS model to investigate the conformational landscape of tau without and with phosphorylations. Characterizing the local compactness of IDPs allows us to highlight how disorder comes in various flavors, as we can define different domains along the tau sequence. We define the IDP’s Statistical Tertiary Organization (STO) as the average spatial arrangements of domains, which constitute an extension of the tertiary structure of folded proteins. We also use IDP-specific metrics to characterize the local curvature and flexibility of tau. Comparing the local flexibilities with T2 relaxation times from NMR experiments, we show how this metric is related to the protein dynamics. A curvature and flexibility pattern in the repeat domains can also be connected to tau binding properties, without having to explicitly model the protein’s interaction partner. Finally, we rediscuss the original paperclip model that describes the spatial organization of tau and how phosphorylations impact it. The resulting changes in the protein intradomain and interdomain interaction pattern allow us to propose experimental setups to test our hypothesis.
"Across (Conformational) Space and (Relaxation) Time", @marienj.bsky.social's latest work, is now out in J. Phys. Chem. B @pubs.acs.org. In this paper, we used coarse-grain simulations with the CALVADOS model to investigate the dynamics of a large IDP.
pubs.acs.org/doi/10.1021/...
19.11.2025 12:14 — 👍 10 🔁 4 💬 1 📌 1
Very excited to present OpenCGChromatin🔥🔥🔥
A new coarse-grained model that probes full chromatin condensates at near-atomistic resolution to reveal the molecular regulation of chromatin structure and phase separation
Brilliantly led by @kieran-russell.bsky.social, with the Rosen and Orozco groups
18.11.2025 15:07 — 👍 48 🔁 15 💬 2 📌 1
An optimized contact map for GōMartini 3 enabling conformational changes in protein assemblies www.biorxiv.org/content/10.1...
16.11.2025 10:36 — 👍 8 🔁 2 💬 0 📌 0
👉Applications will be reviewed on a rolling basis until the #postodc or #phd #position is filled. 📩 Send us your message to explain why you are a great candidate to join the group "Computational Biomolecular Dynamics" of Bert de Groot @mpi-nat.bsky.social 🔗 www.mpinat.mpg.de/degroot #compbiophys
14.11.2025 13:40 — 👍 4 🔁 3 💬 0 📌 1
CECAM - Integrative and Multiscale Modelling Approaches to Illuminate CryoEM/ET data
🔬 CECAM Workshop: “Integrative and Multiscale Modelling Approaches to Illuminate CryoEM/ET Data”
Join us in Toulouse, 24–26 March 2026, to explore how molecular simulations, AI tools, can be integrated to cryo-EM and cryo-ET data.
🧬 Selection: brief CV + motivation letter required.
Apply 👇
01.11.2025 09:47 — 👍 8 🔁 5 💬 0 📌 1
Are you looking for a PhD opportunity? Or would you like to start your postdoc after having earned your doctorate?
Our Department of Theoretical and Computational Biophysics @compbiophys.bsky.social is hiring: ⬇️
29.10.2025 09:05 — 👍 3 🔁 1 💬 0 📌 0
Huge thanks to everyone who was involved: Maxim @maxotubule.bsky.social Helmut @compbiophys.bsky.social and especially Carter @carterjwilson.bsky.social!
27.10.2025 08:00 — 👍 3 🔁 0 💬 0 📌 0
📜 New preprint! We developed PoGö, an algorithm to optimize the essential dynamics of GöMartini proteins based on all-atom simulations: www.biorxiv.org/content/10.1...
27.10.2025 07:54 — 👍 33 🔁 12 💬 2 📌 3
**Job Alert** Exciting postdoc position available: theoretical and experimental cryo-EM studies of flexible biomolecules. Competitive salary, collaborative environment at NYSBC and Flatiron Institute. Please share!! and contact: pcossio@flatironinstitute.org
22.10.2025 14:55 — 👍 14 🔁 18 💬 0 📌 0
Check out our new work, led by talented @janhuemar.bsky.social, where we uncover that pioneer factor Oct4 remodels chromatin for DNA access not by opening it but by exploiting nucleosome breathing and forming clusters 🔥🔥🔥
22.10.2025 05:10 — 👍 11 🔁 9 💬 0 📌 0
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
20.10.2025 11:26 — 👍 91 🔁 37 💬 0 📌 0
Dick is one of the most thoughtful folks in cell biology--this is a must read for anyone interested in microtubules!
15.10.2025 18:23 — 👍 35 🔁 12 💬 0 📌 0

Eye of the mantis shrimp. Bali/Amed with @cecclementi.bsky.social
07.09.2025 07:55 — 👍 11 🔁 1 💬 1 📌 0

Helmut Grubmüller
📢 We are hiring #PhD students and #Postdocs at the
@mpi-nat.bsky.social in
in the Department of Theoretical and Computational #Biophysics in Göttingen 🇩🇪! 🔗 check our current job adverts at www.mpinat.mpg.de/grubmueller and find your project. Submit your #application today! #job
02.09.2025 12:33 — 👍 10 🔁 11 💬 1 📌 0

👉 Open #PhD and #Postdoc positions in #computational #biophysics @mpi-nat.bsky.social in Göttingen 🇩🇪 in the Computational Biomolecular Dynamics group of Bert deGroot for several projects! Applications welcome! 🔗https://www.mpinat.mpg.de/5088717/24-25?c=645962 📝ausschreibung24-25@mpinat.mpg.de
02.09.2025 12:07 — 👍 3 🔁 1 💬 0 📌 0
We are recruiting 2 postdocs to work on bacterial motility using a combination of cryoEM, FLM and MD. An exciting collaboration with Kirsty Wan @micromotility.bsky.social Wolfram Moebius @wolframmoebius.bsky.social and Daniel Kattnig. Please see links in the thread below!
28.08.2025 09:13 — 👍 27 🔁 15 💬 2 📌 2
Coley Research Group - Positions
Positions
#PostDoc opportunity in data-driven #SynChem planning at MIT (Cambridge MA USA) duration: 2 years #cheminformatics #DataScience #CompChem #ChemPostDoc #PostDocJobs #chemsky 🧪
coley.mit.edu/positions
12.08.2025 07:30 — 👍 7 🔁 4 💬 0 📌 0
A few highlights from our new preprint:
On taxol-stabilized microtubules In vitro, wild-type (WT) and phosphor-resistive (AP) tau form well-defined envelopes, while phosphomimetic (E14) tau does not bind cooperatively.
04.08.2025 17:43 — 👍 10 🔁 2 💬 3 📌 0
Intrinsic disorder comes in various flavors ! Using CALVADOS simulations of the Tau protein, we reveal that its disordered dynamics can be interpreted at both residue and domain scales, and that addition of phosphorylations impact both
By @sacquin-mo.bsky.social , Chantal Prévost and I
25.07.2025 06:59 — 👍 18 🔁 6 💬 1 📌 1
Our paper on:
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
22.07.2025 05:58 — 👍 41 🔁 13 💬 1 📌 1
Arriën & Giulio's paper on
A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social
17.07.2025 08:54 — 👍 13 🔁 5 💬 0 📌 0
4th year PhD Student, Collepardo Lab, University of Cambridge
Associate Professor at MIT Chemistry; Enthusiastic about chromatin, biophysics, molecular dynamics, and AI.
Predoctoral Research Assistant at the Llorca lab @cniostopcancer.bsky.social working on mTORC1 signaling and cryo-EM ❄️🔬. Intrigued by the structural mechanisms of protein assemblies involved in endomembrane signaling and trafficking 🧪.
Professor of Microbial Biophysics, University of Warwick. Interested in all things bacterial cell envelope, cell biophysics and advanced light microscopy. https://holdenlab.github.io/ Twitter (inactive): @seamus_holden
Post doctoral researcher at University of Illinois Chicago, Pharmaceutical science department.
Independent PostDoc Fellow at HITS Heidelberg (@hitsters.bsky.social) working on chemical space exploration for nanomedicine using multi-scale #compchem methods
American Cancer Society Postdoctoral fellow and structural biologist in the @briankelch.bsky.social lab at UMass Chan Medical school. I study ATPases and DNA replication/repair. Phage and MD simulations were my first scientific love. he/him/his
Computational biologist @ Roche | Views my own
Die DFG ist die größte Forschungsförderorganisation und die Selbstverwaltungsorganisation der Wissenschaft in Deutschland.
www.dfg.de
Impressum: https://www.dfg.de/de/service/kontakt/impressum
Social Media-Netiquette: https://www.dfg.de/de/service/presse
PhD student in Computational Structural Biology @Utrecht University
ML for structural bioinformatics
pharmacist, PhD, researcher, medicinal chemist, drug discovery, neglected diseases, onehealth, Leismania
Explore groundbreaking news and research from PNAS, one of the world's most-cited scientific journals. Discover its sibling journal, @pnasnexus.org, both official journals of the National Academy of Sciences. Visit www.pnas.org for more info.
Interests in ML and social aspects of tech.
Building For You feed: https://bsky.app/profile/spacecowboy17.bsky.social/feed/for-you
Hobby project: linklonk.com
Professor at the NYU School of Medicine (https://yanailab.org/). Co-founder and Director of the Night Science Institute (https://night-science.org/). Co-host of the 'Night Science Podcast' https://podcasts.apple.com/us/podcast/night-science/id1563415749
Genomics, Machine Learning, Statistics, Big Data and Football (Soccer, GGMU)
machine learning and functional/statistical genetics. Associate Prof @Columbia and Core Faculty @nygenome. he/him/his. https://daklab.github.io/
Scientist, Assistant Professor at MIT biology, #FirstGen
Computational structural biologist @Fox Chase Cancer Center. Gay, out & proud since 1986. I support transgender & nonbinary people. Living with CLL/SLL. Video about my LGBTQ+ life in science: https://tinyurl.com/45srzjdw. Views my own, not my employer's.
Open source methods for #metagenomics with @martinsteinegger.bsky.social Seoul National University 🇰🇷 | former Söding Lab MPI-NAT 🇩🇪 | http://mstdn.science/@milotmirdita