Photoproximity labeling of c-Myc reveals SLK as a cancer specific co-regulator https://www.biorxiv.org/content/10.1101/2025.10.02.680136v1
05.10.2025 03:49 — 👍 1 🔁 1 💬 0 📌 0
Happy to share the finalized version of this work out in @angewandtechemie.bsky.social.
onlinelibrary.wiley.com/doi/10.1002/...
12.09.2025 17:53 — 👍 10 🔁 2 💬 0 📌 0
Eragon series is good. Bartimaeus also good.
15.07.2025 00:37 — 👍 1 🔁 0 💬 0 📌 0


Crazy new pre-print from TSRI: fix whole mice, clear them transparent, click on fluorophores, & use light sheet microscopy to visualize where drugs bind throughout the body. Find ibrutinib-yne in low BTK heart & blood vessels, correlates with cardiac side effects.
www.biorxiv.org/content/10.1...
05.04.2025 13:39 — 👍 21 🔁 7 💬 1 📌 0

Engineered Proteins and Chemical Tools to Probe the Cell Surface Proteome
The cell surface proteome, or surfaceome, is the hub for cells to interact and communicate with the outside world. Many disease-associated changes are hard-wired within the surfaceome, yet approved drugs target less than 50 cell surface proteins. In the past decade, the proteomics community has made significant strides in developing new technologies tailored for studying the surfaceome in all its complexity. In this review, we first dive into the unique characteristics and functions of the surfaceome, emphasizing the necessity for specialized labeling, enrichment, and proteomic approaches. An overview of surfaceomics methods is provided, detailing techniques to measure changes in protein expression and how this leads to novel target discovery. Next, we highlight advances in proximity labeling proteomics (PLP), showcasing how various enzymatic and photoaffinity proximity labeling techniques can map protein–protein interactions and membrane protein complexes on the cell surface. We then review the role of extracellular post-translational modifications, focusing on cell surface glycosylation, proteolytic remodeling, and the secretome. Finally, we discuss methods for identifying tumor-specific peptide MHC complexes and how they have shaped therapeutic development. This emerging field of neo-protein epitopes is constantly evolving, where targets are identified at the proteome level and encompass defined disease-associated PTMs, complexes, and dysregulated cellular and tissue locations. Given the functional importance of the surfaceome for biology and therapy, we view surfaceomics as a critical piece of this quest for neo-epitope target discovery.
Excited to share that our cell surface proteome review is now online on Chemical Reviews! 🥰 We highlight recent advances of techniques mapping cell surface protein expression, protein-protein interactions, extracellular PTMs and MHC complexes. @jimwellsucsf.bsky.social pubs.acs.org/doi/10.1021/...
04.04.2025 20:19 — 👍 49 🔁 10 💬 2 📌 1
Congrats!!!
21.03.2025 13:10 — 👍 2 🔁 0 💬 1 📌 0
Absolutely honored to be selected 2025 CAS Future Leaders! Congrats to all the cohort and I look forward to join you in upcoming events! 🥳
20.03.2025 17:23 — 👍 11 🔁 2 💬 2 📌 0
Thanks Neel!! Following behind you and Tom’s foundation last year.
20.03.2025 18:33 — 👍 2 🔁 0 💬 0 📌 0
New Preprint from my group! We develop a systems level proximity labeling platform to study MOA of transcriptionally active drugs. In collaboration with the Rumbaugh lab, we employ this to deorphan a candidate ligand to treat neurodevelopmental disorders. www.biorxiv.org/content/10.1...
18.03.2025 20:29 — 👍 1 🔁 0 💬 0 📌 0

Deep-Red and Ultrafast Photocatalytic Proximity Labeling Empowered In Situ Dissection of Tumor-Immune Interactions in Primary Tissues
Immunotherapy efficacy in solid tumors varies greatly, influenced by the tumor microenvironment (TME) and the dynamic tumor-immune interactions within it. Decoding these interactions in situ with minimal interference with native tissue architecture and delicate immune responses is critical for understanding tumor progression and optimizing therapeutic strategies. Here, we introduce CAT-Tissue, a novel deep-red photocatalytic proximity labeling method that enables ultrafast, high-resolution profiling of tumor-immune interactions in primary tissues. By leveraging nanobody-Chlorin e6 as the photocatalyst and biotin-aniline as the probe, CAT-Tissue enabled the rapid and comprehensive detection of various tumor-immune interactions in both coculture systems and primary tumor sections. Coupled with bulk RNA-sequencing, CAT-Tissue revealed distinct gene expression patterns between tumor-neighboring and tumor-distal lymphocytes, highlighting the recognition and immune responses of tumor-neighboring CD8+ T cells, which exhibited activated, effector, and exhausted phenotypes. By leveraging a deep-red photocatalytic proximity cell labeling strategy with excellent tissue penetration and biocompatibility, CAT-Tissue offers a nongenetically encoded platform with high sensitivity and spatiotemporal controllability for rapid profiling tumor-immune interactions within complex tissue environments in situ, which may advance our understanding of tumor immunology and guide the development of more effective immunotherapies.
See also this awesome work from Peng Chen and Xinyuan Fan who published a similar method earlier this week. pubs.acs.org/doi/10.1021/...
07.03.2025 20:05 — 👍 1 🔁 0 💬 0 📌 0

Multiprobe Photoproximity Labeling of the EGFR Interactome in Glioblastoma Using Red-Light
Photocatalytic proximity labeling has emerged as a valuable technique for studying interactions between biomolecules in a cellular context, providing precise spatiotemporal control over protein labeling. One significant advantage of these methods is their modularity, allowing the use of a single photocatalyst with different reactive probes to expand interactome coverage and capture diverse protein interactions. Despite these advances, fewer methods have been developed using red-light excitation, limiting the use of photoproximity labeling in more complex media such as tissues and animal models. Herein, we develop a platform for proximity labeling under red-light excitation, utilizing a single catalyst and two distinct probe types. We first design a carbene based labeling system that utilizes sulfonium diazo probes. This system is successfully applied on A549 cells to capture the interactome of epidermal growth factor receptor (EGFR) using a Cetuximab-Chlorin e6 conjugate. Benchmarking against established techniques indicates that this approach performs comparably to leading carbene-based proximity labeling methods. Next, we leverage the strong singlet oxygen generation (SOG) ability of Chlorin e6 to establish an alternative labeling system using aniline and hydrazide probes. EGFR directed chemoproteomics experiments reveal significant overlap with the carbene system, with the carbene approach capturing a subset of interactions identified by the SOG system. Finally, we deploy our approach for the characterization of EGFR in resected human glioblastoma (GBM) tissue samples removed from distinct locations in the same tumor, representing the tumor’s infiltrating edge and its viable center, identifying several GBM specific interacting proteins that may serve as a launch point for future therapeutic campaigns.
Our first paper is out today in @jamchemsoc.bsky.social!
This is the beginning of a long journey for us, to study heterogeneity in GBM patient samples.
First step was to figure out some chemistry to enable us to look at this challenging problem.
pubs.acs.org/doi/10.1021/...
07.03.2025 19:36 — 👍 6 🔁 3 💬 1 📌 1
There is no need for different names for different flavors of display item in journals. Figures, Schemes, Tables, etc. can all just be called Figures and nothing is lost
07.03.2025 03:10 — 👍 35 🔁 4 💬 6 📌 0
Cell Surface Map
Cell Surface Map - Distribution and organization data on immune cell surface proteins
Check out our web tool for searching for interactors of your favorite cell surface protein, developed by undergrad Pinyu Liao and postdoc postdoc Brendan Floyd @stanford-chemh.bsky.social
cellsurfacemap.org
21.02.2025 01:56 — 👍 155 🔁 37 💬 0 📌 1
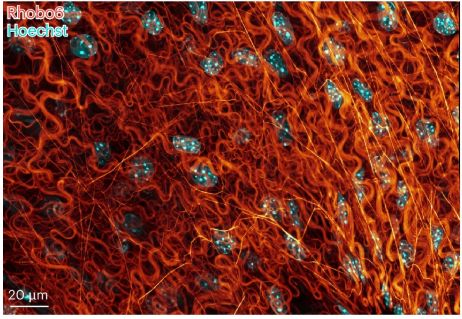
"Imagine a DAPI-like stain, but for the extracellular matrix." That's basically how this work was pitched to me by Kayvon and Antonio a year or so ago. Now the final product really delivers. Read about their versatile label for ECM in living tissues here: www.nature.com/articles/s41...
06.02.2025 16:02 — 👍 354 🔁 104 💬 9 📌 7

Applications for the 2025 Bioorganic GRC are now live! Here is the link where you can check out the current line-up:
grc.org/bioorganic-c...
16.11.2024 22:23 — 👍 63 🔁 30 💬 2 📌 9
Come hang with me and @zhilindseylin.bsky.social at the 2025 Bioorganic Chemistry GRS! It’ll be a weekend full of excellent science and even more excellent people. The GRS is one of my favorite weekends of the year and we’re looking forward to another seminar full of amazing, trainee-led science!
16.11.2024 22:29 — 👍 9 🔁 5 💬 1 📌 1
Postdoc @ Lacra Bintu Lab
Scientist interested in 3’UTRs and mRNA-based translation environments. We study how mRNAs regulate IDR conformations and protein functions. MSKCC in NYC
I am an Assistant Professor of Cancer Biology at Loyola University Chicago Stritch School of Medicine (www.fanninglab.com). The Fanning lab studies how allostery governs estrogen receptor-dependent transcriptional programs in breast cancer.
Asst Prof of Pharmaceutical Sciences #UCIrvine | Studying #microprotein and #peptide biology in cancer and metabolism | #FirstGen | #LAnative | 🇵🇸🇲🇽-🇺🇸 | https://faculty.sites.uci.edu/martinezlab/
Assistant Professor, UW-Madison Chemistry. Group website: http://martellgroup.chem.wisc.edu.
Chief Editor of Nature Biomedical Engineering. Ph.D. Previously at Nature Methods. @rita_strack on the place formerly known as twitter.
'This Is My Glasgow' is a photographic project from Colin M. Drysdale, author of 'Glasgow Uncovered: 18 Walks Through its Past, Present and Future.' All images are my own.
Organic chemist, currently postdoc in the Shenvi lab at Scripps Research.
Chemical Biologist at Penn Medicine. Burslemlab.com
Confused Brit in the US. He/him
Editor at Science Magazine, shepherding chemistry papers; views here are my own; he/him
Medchem PhD student in Farnaby group @CeTPD Dundee focusing on CNS diseases and target protein degradation
Assistant Professor at Columbia Chemistry. PI of a chemical biology lab full of awesome people. Internal conflicts: Chemist or biologist? Kinase or phosphatase?
Lab website: https://shahlab.wixsite.com/home
ORCiD: 0000-0002-1186-0626
Waitress turned Congresswoman for the Bronx and Queens. Grassroots elected, small-dollar supported. A better world is possible.
ocasiocortez.com
Professor and Director, Dept. of Biophysics & Biophysical Chemistry
Johns Hopkins School of Medicine, structural biologist, lover of chromatin and ubiquitin, fan of the active voice.
Assistant professor at UF Scripps. Aiming to understand and disrupt cancer ecosystem.
Chemist. PI of the Gómez-Suárez lab (radical.uni-wuppertal.de) and scientific staff of the Kirsch group (oc-sfk.uni-wuppertal.de) at the Bergische Universität Wuppertal.
PhD Student @GloriusGroup working on EnT Catalysis
Professor @ Scripps Research | Department of Chemistry | chemical biology & chemical proteomics
Senior Scientist @ BMS Discovery Chemistry Hobby Enthusiast: Running, Crocheting, Knitting, Crossstitch, Board Games, Hiking, Camping
🥼👟🧶🪡🧵🏞🏕











