Scripps Research team identifies sugar molecules that trigger placental formation
Using proximity labeling and a derivatized galectin-3, scientists in the lab of Professor Mia Huang mapped glycan–protein interactions that drive placental cell fusion, published in @pnas.org, offering new insight into placental development.
08.11.2025 00:25 — 👍 9 🔁 7 💬 0 📌 0
Apply - Interfolio
{{$ctrl.$state.data.pageTitle}} - Apply - Interfolio
During these uncertain times, I’m very happy to see that my institution, @scripps.edu has an open tenure-track Assistant Professor position. Any field in Chemistry or Biology is welcome. I’d especially love to see fellow neuroscientists apply. Please repost!
apply.interfolio.com/174756
14.10.2025 17:35 — 👍 130 🔁 95 💬 2 📌 5

Proteome-Wide Discovery of Degradable Proteins Using Bifunctional Molecules
Targeted protein degradation (TPD) is an emergent therapeutic strategy with the potential to circumvent challenges associated with targets unamenable to conventional pharmacological inhibition. Among ...
Happy to share the final version of this work out in ACS CS. Inspired by ‘binding-focused’ chemoproteomic methods, we developed a ‘function-focused’ strategy to agnostically identify degradable proteins.
This was a big team effort led by
@inesforrest.bsky.social
and in collaboration with AbbVie.
21.10.2025 15:55 — 👍 37 🔁 9 💬 0 📌 1

Discovery of a tau-aggregate clearing compound that covalently targets P4HB
The improper folding and aggregation of tau are linked to several neurodegenerative diseases affecting millions worldwide. However, the pathogenesis o…
Check out our recent collaboration with @abbvie.bsky.social in @cp-cellchembiol.bsky.social. We identifed compounds that potently clear tau aggregates. After chemoproteomics and functional studies, we found it covalently targets ER protein P4HB (after metabolic activation).
06.10.2025 17:01 — 👍 8 🔁 0 💬 1 📌 0
Happy to share the finalized version of this work out in @angewandtechemie.bsky.social.
onlinelibrary.wiley.com/doi/10.1002/...
12.09.2025 17:53 — 👍 11 🔁 2 💬 0 📌 0

Benjamin Cravatt awarded Tetrahedron Prize for Creativity - Scripps Research Magazine
Benjamin Cravatt, the Norton B. Gilula Chair in Biology and Chemistry at Scripps Research, will receive the 2025 Tetrahedron Prize for Creativity in Bioorganic and Medicinal Chemistry. Established in ...
Congrats to Benjamin Cravatt on receiving the 2025 Tetrahedron Prize for Creativity in Bioorganic & Medicinal Chemistry from @elsevierconnect.bsky.social, honoring his pioneering activity-based protein profiling (ABPP) to study and target disease-relevant proteins.
13.08.2025 00:13 — 👍 52 🔁 8 💬 0 📌 0
Beyond thrilled to share that my PhD work is now available as a preprint! 🥳 A huge thank you to my amazing colleagues, mentors and collaborators at @scripps.edu and @abbvie.bsky.social who supported and contributed to this project!
24.03.2025 07:34 — 👍 9 🔁 1 💬 0 📌 0
😎
02.04.2025 07:30 — 👍 1 🔁 0 💬 0 📌 0


The sun is shining in London for the first day of #ABPP2025 ! Really excited to welcome everyone to the Crick later with an incredible lineup for the first session @abpp2025.bsky.social
30.03.2025 10:01 — 👍 10 🔁 1 💬 2 📌 0
Looking forward to catching up!
28.03.2025 01:57 — 👍 1 🔁 0 💬 0 📌 0
We designed a library of function-biased, target agnostic bifunctional molecules and profiled them proteome wide for their ability to degrade proteins. We believe this study provides a proof of concept to expand PROTAC strategies beyond targets with well-established ligands.
23.03.2025 07:16 — 👍 3 🔁 0 💬 0 📌 0
Congratulations!!!
22.03.2025 21:27 — 👍 1 🔁 0 💬 0 📌 0
I’m a standing member on the Chemical Biology and Probes (CBP) NIH study section that was supposed to meet tomorrow. It just got postponed this afternoon with a date yet to be determined….
19.02.2025 20:22 — 👍 23 🔁 12 💬 3 📌 1
🚨 Chemical Biology & Probes study section (formerly SBCB, one of two NIH panels that reviews chemistry #chemsky 🧪) was abruptly POSTPONED w/no specific plans for rescheduling, less than 24h before start.
If this affects you call your reps & senators, talk to local news, make your voice heard 📢 👩🔬
19.02.2025 19:55 — 👍 35 🔁 27 💬 4 📌 5
Congrats Brian and team!!!
12.02.2025 17:40 — 👍 3 🔁 0 💬 1 📌 0
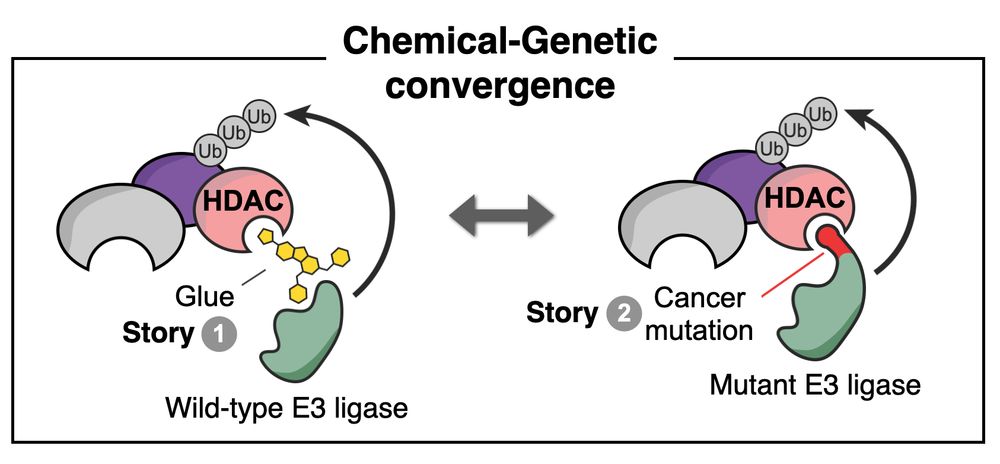
Today in @nature.com we share our back-to-back stories with Ning Zheng’s lab revealing chemical-genetic convergence between a molecular glue degrader & E3 ligase cancer mutations. 1/5
12.02.2025 16:20 — 👍 87 🔁 40 💬 5 📌 5
Check out our recent efforts in chemrxiv to expand the chemical tractability of the proteome through NP-inspired photoaffinity probes
chemrxiv.org/engage/chemr...
05.02.2025 07:46 — 👍 14 🔁 3 💬 0 📌 1

Insightful perspective by the group of Ben Cravatt with first author @xiaoyuzhang.bsky.social in @nchembio.bsky.social. They discuss reasons, why expanding the repertoire of E3s for TPD has converged on few "frequent hitters" and how additional E3 targets could be identified. doi.org/10.1038/s415...
29.01.2025 06:30 — 👍 4 🔁 1 💬 0 📌 0
Congrats Ben!!!
08.01.2025 15:05 — 👍 2 🔁 0 💬 0 📌 0
Chancellor's Distinguished Professor of Physics, UCSD. Interested in everything.
Theoretical physicist researching condensed & living matter, fluid mechanics, statistical mechanics. Bad at staying in my lane.
https://guava.physics.ucsd.edu/~nigel/
Assistant Professor at UF-Scripps and the College of Pharmacy, UF
Chemical proteomics, chemical biology, ligand discovery
Postdoc @scripps.edu interested in protein-protein interactions and all things undruggable.
Structural biologist and biochemist. CNRS researcher at CBM Orléans @cbm-upr4301.bsky.social. Interested in protein modifications & interactions. Also husband, dad of 2, friend, ☧. Personal website: msuskiewicz.github.io
Organic chemist with a weird accent from New Jersey
Science, glycobiology, mass spectrometry, coffee. Assoc. professor @unimelb. Former ARC future fellow. he/his
Chemical Biologist. Loves probes, hates TB.
Assistant prof at Stanford. Interested in aneuploidy, mitotic kinases, cancer therapeutics, and drug development. Co-founder x2.
Professor and Head of the Division of Molecular Therapeutics in MCB, UC Berkeley
Founder, Nurix and Lyterian Therapeutics
Excited about stress responses, ubiquitin, proteasome, targeted protein degradation, drug discovery
chemical biology enthusiast & strategist; I make impossible things possible - all before breakfast!
Chemist at HHMI's Janelia Research Campus. Passionate about designing, building, and giving away fluorescent dyes to illuminate biological systems. Striving to be positive about all things chemistry (except ChemDraw).
ORCID: 0000-0002-0789-6343
Assistant Professor, UW-Madison Chemistry. Group website: http://martellgroup.chem.wisc.edu.
Trying to change the world, 1 reaction at a time at #MerckChemistry. Fellow of the Royal Society of Canada. Cohost of the Pharm to Table Podcast. #ChemistsWhoCook👨🍳 #ChemistsWhoGolf🏌️♂️
I write about medicinal and computational chemistry and drug discovery with a sprinkling of history. Blog at medchemash.substack.com.
Immunologist. T cell exhaustion, immunotherapy, Immune Health.
Husband to Pam Sharma. Immunologist. Nobel Laureate.
Expert recommendations and info about #ChemicalProbes, particularly small molecule inhibitors, and their use in biomedical research and #DrugDiscovery.