Preprint from the lab🚨
Have you ever engineered proteins to be more stable and were unhappy about your predictor's success rate? We got you covered with BoostMut!
Great work led by @kerlenkorbeld.bsky.social now online at www.biorxiv.org/content/10.1...
A thread 🧵
07.05.2025 17:04 — 👍 43 🔁 8 💬 1 📌 0
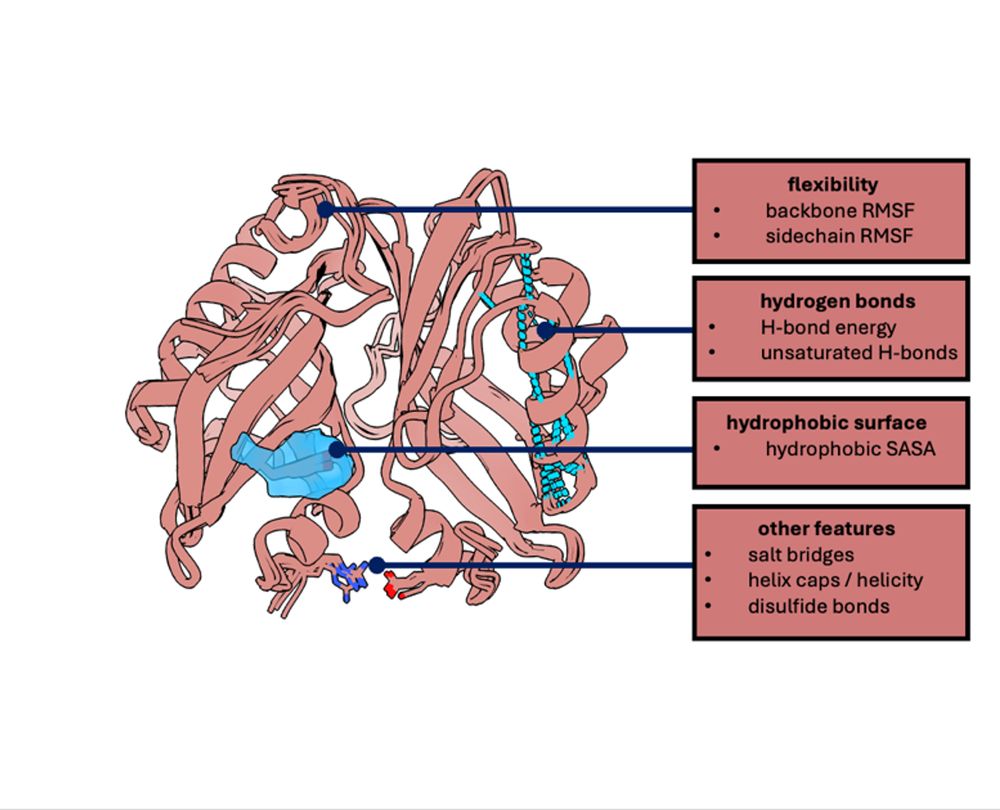
Inverse folding NNs are better at predicting changes to a mutated protein's equilibrium dynamics, relative to WT, than protein folding NNs (BioEmu & AF-Cluster). This was shown on an enzyme that is more enzymatically promiscuous in the open state.
12.05.2025 06:27 — 👍 23 🔁 7 💬 0 📌 0

De novo design of porphyrin-containing proteins as efficient and stereoselective catalysts
De novo design of protein catalysts with high efficiency and stereoselectivity provides an attractive approach toward the design of environmentally benign catalysts. Here, we design proteins that inco...
Designing enzymes w/ a combination of structure-based sequence design (LigandMPNN) and directed evolution. The latter step discovers unconventional mutations like a mid-helical proline, and subsequent MD simulations show an increasingly rigidified active site
15.05.2025 13:25 — 👍 24 🔁 6 💬 0 📌 0
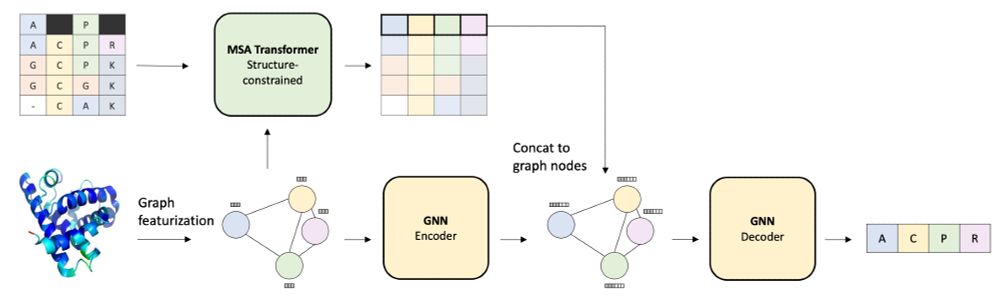
“A joint embedding of protein sequence and structure enables robust variant effect predictions”
Combines a structure GNN w/ MSA-transformer, partly by “only allowing attention between positions in the MSA that are proximal in” 3D space
www.biorxiv.org/content/10.1...
github.com/KULL-Centre/...
17.12.2023 13:09 — 👍 5 🔁 2 💬 0 📌 0

Altae-Tran, Kannan, Koonin, Zhang + al. reports @Science a new protein clustering algorithm + discovery of rare and previously unknown CRISPR-Cas systems - an untapped trove of diverse biochemical activities linked to RNA-guided mechanisms w/ potential for biotechnology development bit.ly/adi1910
25.11.2023 19:17 — 👍 20 🔁 8 💬 1 📌 0
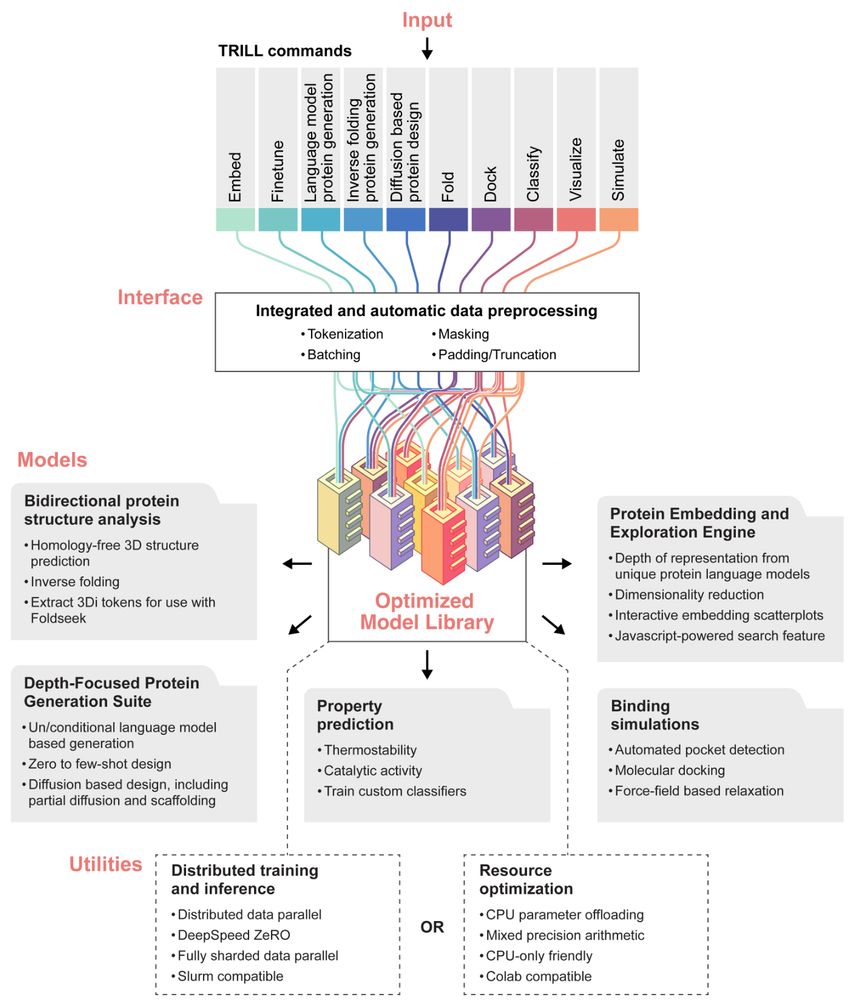
“TRILL: Orchestrating Modular Deep-Learning Workflows for Democratized, Scalable Protein Analysis and Engineering” has been updated 🧪🧶🧬
Provides a framework to integrate PLMs, backbone diffusion, docking, inverse folding, etc
www.biorxiv.org/content/10.1...
trill.readthedocs.io/en/latest/ho...
11.11.2023 11:51 — 👍 13 🔁 4 💬 0 📌 0

“Design of efficient artificial enzymes using crystallographically-enhanced conformational sampling” 🧪🧶🧬
Engineering a Kemp eliminase w/ 250-fold increase in enzymatic activity using an experimental structural ensemble and molecular dynamics-based filtering
www.biorxiv.org/content/10.1...
03.11.2023 06:04 — 👍 9 🔁 3 💬 1 📌 0

“The genetic architecture of protein stability” 🧪🧶🧬
An additive fitness prediction model trained on ≤2 mutations predicts fitness effects of 3-13 substitutions with R2=0.5
www.biorxiv.org/content/10.1...
28.10.2023 05:49 — 👍 8 🔁 3 💬 0 📌 0

Enzymatic synthesis and nanopore sequencing of 12-letter supernumerary DNA - Nature Communications
Unnatural base pairing xenonucleic acids (XNAs) can be used to expand life’s alphabet beyond ATGC. Here, authors show strategies for enzymatic synthesis and next-generation nanopore sequencing of XN...
Here, we report on strategies for both writing and reading DNA with expanded alphabets composed of up to 12 letters (A, T, G, C, B, S, P, Z, X, K, J, V).
www.nature.com/articles/s41...
28.10.2023 04:00 — 👍 0 🔁 0 💬 0 📌 0

“OPUS-Rota5: A Highly Accurate Protein Side-chain Modeling Method with 3D-Unet and RotaFormer” 🧪🧶🧬
Better prediction quality than AlphaFold2 on CASP15 targets
www.biorxiv.org/content/10.1...
github.com/OPUS-MaLab/o...
21.10.2023 05:53 — 👍 7 🔁 1 💬 0 📌 0

“Predicting a Protein's Stability under a Million Mutations” 🧪🧶🧬
Structure-based stability prediction of single- and double-point mutants
arxiv.org/abs/2310.12979
github.com/jozhang97/Mu...
20.10.2023 05:09 — 👍 6 🔁 3 💬 0 📌 0
A working microbiologist ... interested in #oxidoreductases, #flavoproteins, #biocatalysis and #degradation of #xenobiotics by #actinomycetes, #naturalproduct, #biotransformation and #enzyme #catalysis
Hobby #ChemistsWhoCook
Australian microbial biochemist, synthetic biologist and protein engineer. CSO at ULUU. Interests in biotech solutions to pollution, resource scarcity, and sustainable manufacture.
We study enzyme structure and dynamics using biophysical and biochemical tools at Cornell University 🧪 Posts made by students in the Ando Lab
PI: @nozomi-ando.bsky.social
Tenure-track Assistant Professor at TUM #Biocatalysis #Enzymes #EnzymeImmobilization #IDPs #ProteinStability #NaturalProducts
Chemical Biology Lab - Interests in Protein Chemistry; Post-translational modifications, especially those that occur on the polypeptide backbone; p53 signalling; molecular ageing.
An open access journal indexed in: #ESCI #PMC #DOAJ #Scopus (#CiteScore2023: 3.9) etc. #Synbio #biodesign #SyntheticBiology #openaccess
A gold open-access journal focused on innovative chemical biology tools and their application in exploring and manipulating molecular biological processes.
🌐 Website: rsc.li/rscchembio Published by @rsc.org
Cutting-edge research, news, commentary, and visuals from the Science family of journals. https://www.science.org
RCSB PDB (RCSB.org) promotes a structural view of biology. Funded by NSF, NCI, NIAID, NIGMS, NIH, and DOE
Developing macromolecular structure resources and tools for life science.
- Founding partner of the wwPDB.
- Manage the community-led PDBe-KB resource and the AlphaFold Protein Structure Database, a collaboration with Google DeepMind.
- Part of EMBL-EBI
Enthusiastic about Biocatalysis, Evolution and Extremophiles. Loves microbes, microengineering and meditation. @KITKarlsruhe
@NiemeyerLab. Views are my own.
Photocatalysis, Supramolecular Catalysis, Total Synthesis, and C-H Activation (student run account)
Organic Chemistry & Biomimetic Catalysis @ Saarland University and Helmholtz Institute for Pharmaceutical Research Saarland
A catalysis jack of all trades. I support Chemistry for Good.
Executive Editor of PCCP, Green Chemistry and Faraday Discussions,
on behalf of the @roysocchem.bsky.social
Marie Skłodowska-Curie Research Fellow in Biocatalysis at TU Delft|Likes enzymes, space, and tea.
Chemistry professor at Missouri S&T, electrochemist, editor-in-chief of the ACS Au journals
Penn Integrates Knowledge Professor, Natural Product Aficionado, Photopharmacologist, Fox Terrierist, Book Lover, and Unfulfilled Architect. #firstgen
Prof for chemical biology at ETH Zurich
Interested in: genetic code expansion - chembio tools - bioorthogonal chemistries - PTMs - Ub in all its shades - protein engineering














