 28.09.2025 21:50 — 👍 3 🔁 4 💬 0 📌 0
28.09.2025 21:50 — 👍 3 🔁 4 💬 0 📌 0
@charliebell.bsky.social
Senior post-doc in the Faulkner lab ARC DECRA Fellow Interested in how gene regulatory mechanisms contribute to biological complexity
@charliebell.bsky.social
Senior post-doc in the Faulkner lab ARC DECRA Fellow Interested in how gene regulatory mechanisms contribute to biological complexity
18/ At large genomic distance, long encounters (=the important ones) are only due to loop extrusion; but at short genomic distance they can also be due to random collisions. This is why enhancers are no sensitive to loss of extrusion when they are close, but sensitive when they are far!
24.09.2025 21:45 — 👍 2 🔁 1 💬 1 📌 0Really excited to share our latest work led by @mattiaubertini.bsky.social and @nesslfy.bsky.social: we report that cohesin loop extrusion creates rare but long-lived encounters between genomic sequences which underlie efficient enhancer-promoter communication.
www.biorxiv.org/content/10.1...
A🧵👇

We are looking for a student to continue our work on chromatin evolution:
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
The project with @seanamontgomery.bsky.social will focus on chromatin state readers across eukaryotes.
More info: recruitment.crg.eu/content/jobs...
New preprint from the lab 🎉 on the evolution of gene regulatory network complexity, where we tested whether sexual recombination promotes more complex GRNs using biochemically-inspired evolutionary simulations 🧐. Bluetorial from 1st author @chapelmadison.bsky.social 👇
04.09.2025 00:37 — 👍 6 🔁 2 💬 0 📌 0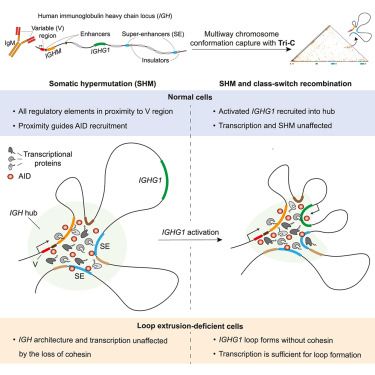
Online Now: Regulation of somatic hypermutation by higher-order chromatin structure Online now:
27.06.2025 22:57 — 👍 19 🔁 7 💬 0 📌 0
So cool, a novel archaeon with a super small genome and no metabolic genes!
"An unprecedented level of metabolic dependence on a host, a condition that challenges the functional distinctions between minimal cellular life and viruses."
www.biorxiv.org/content/10.1...
Many cells during development are exposed to caspase activation and yet survive. Why some die and not others ? We found out that the memory of previous caspase activation bias significantly later death comitment and bias death distribution and single cell decision
www.biorxiv.org/content/10.1...
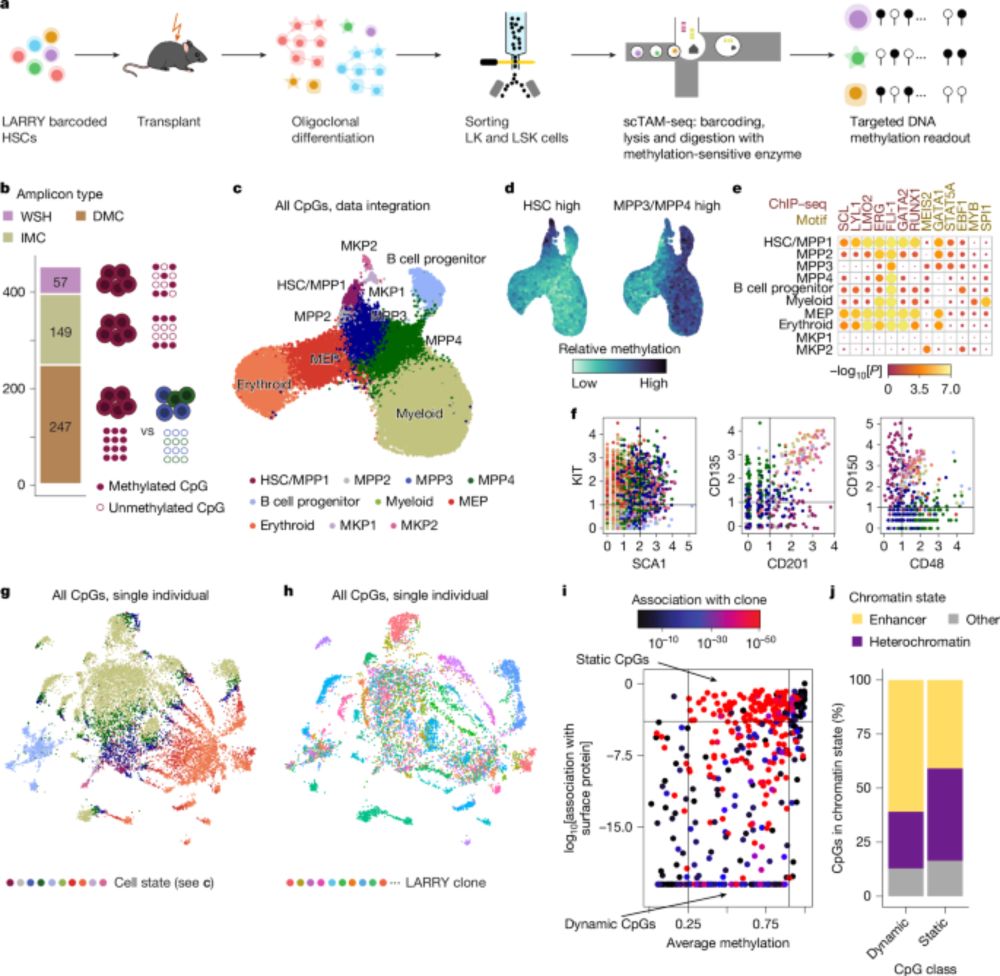
Out @nature.com: Clonal tracing with somatic epimutations
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵

Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
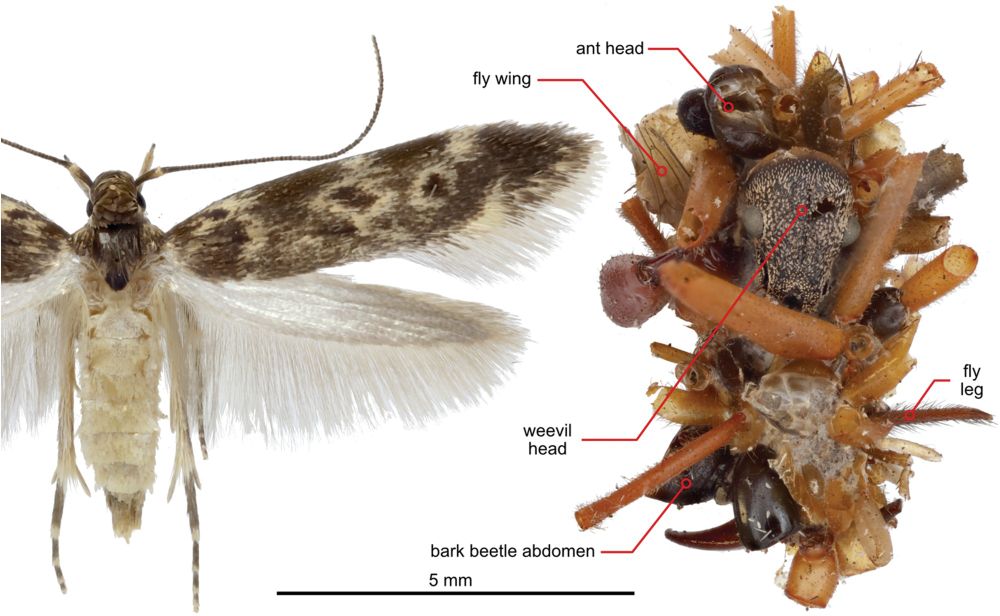

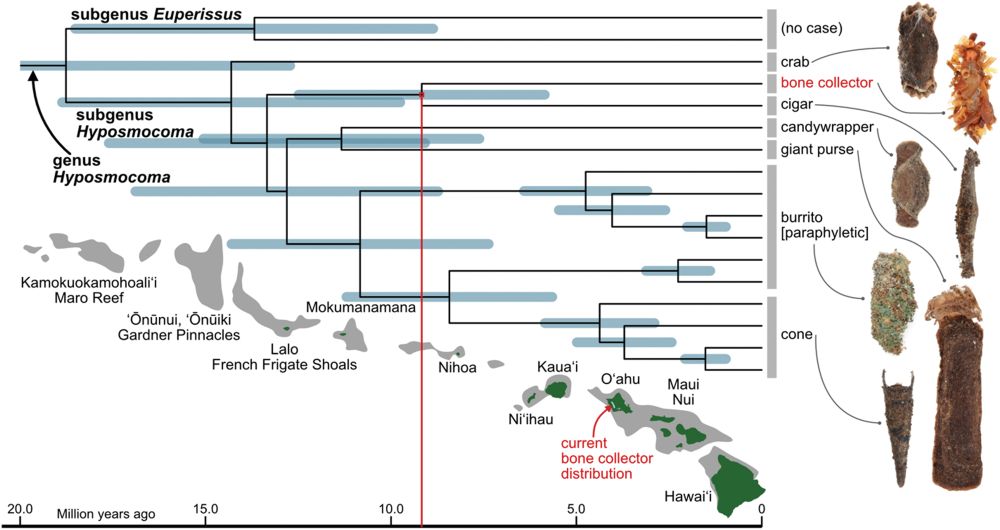
🐛🕷️🕸️ @science.org Hawaiian caterpillar patrols spiderwebs camouflaged in insect prey’s body parts | Science www.science.org/doi/10.1126/... #entomology
24.04.2025 18:41 — 👍 19 🔁 5 💬 0 📌 0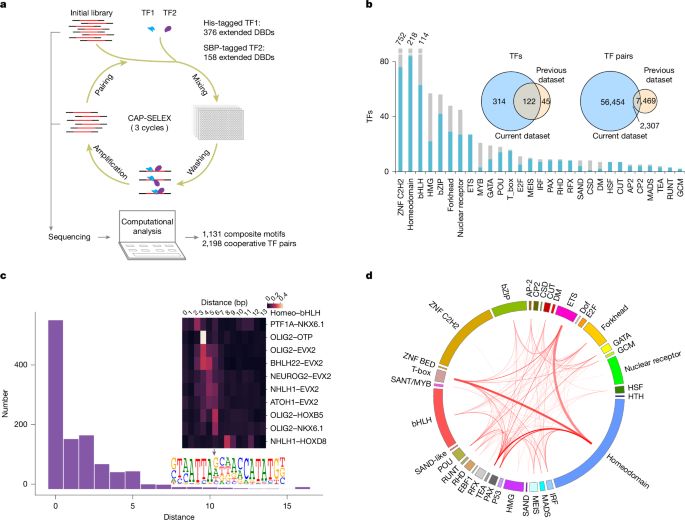
Nature research paper: DNA-guided transcription factor interactions extend human gene regulatory code
https://go.nature.com/4j13XRc
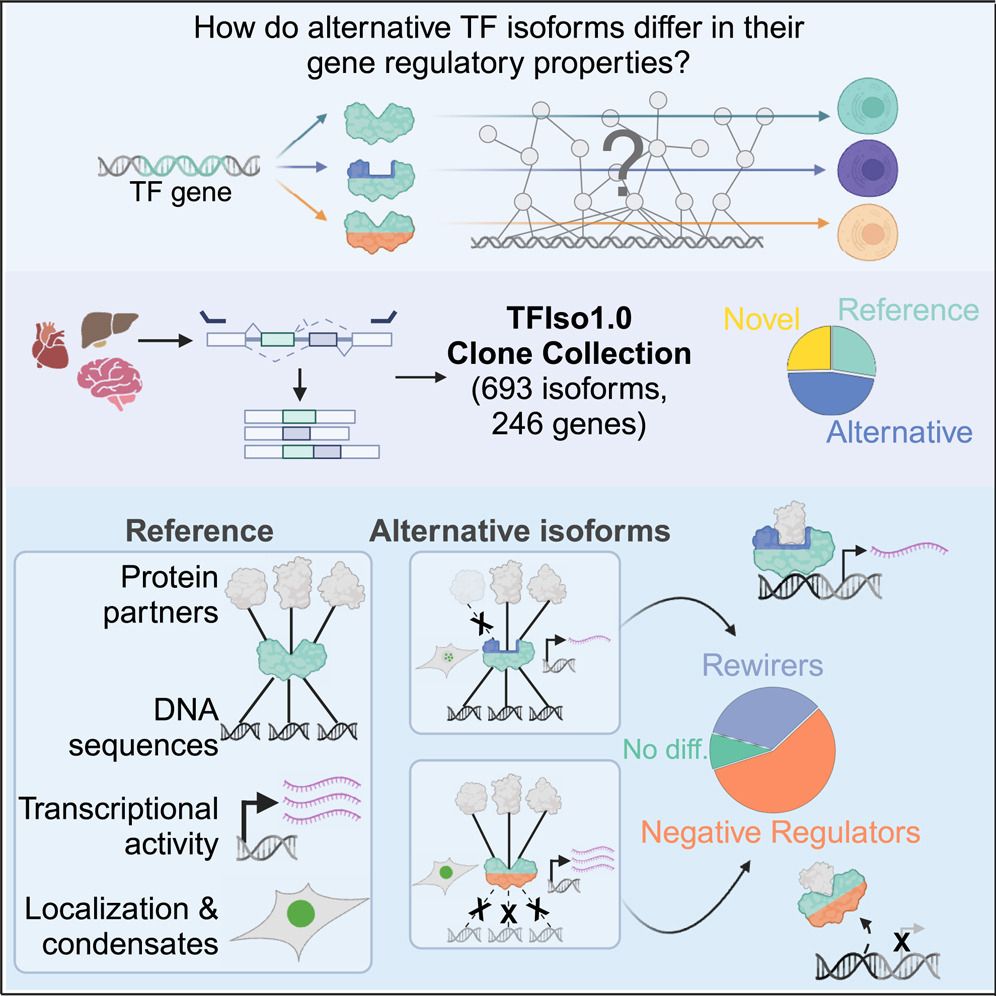
our work on the molecular differences between transcription factor isoforms is out now in Molecular Cell!
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
1/ H3K27me3 mimicry has repeatedly emerged through evolution, but what's the physiological relevance?
We show that JARID2 and PALI1 mimic H3K27me3 to antagonise PRC2 in vivo and restrict the spread of Polycomb domains.
🧵
www.biorxiv.org/content/10.1...
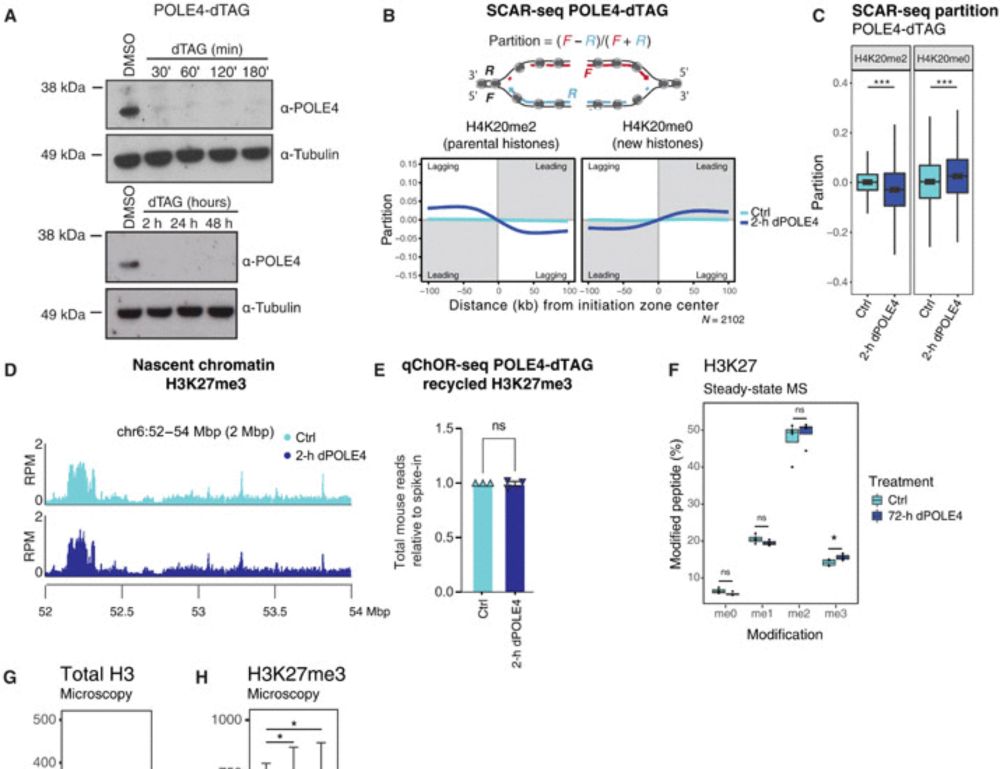
𝐇𝐨𝐰 𝐝𝐨 𝐜𝐞𝐥𝐥𝐬 𝐫𝐞𝐦𝐞𝐦𝐛𝐞𝐫 𝐰𝐡𝐨 𝐭𝐡𝐞𝐲 𝐚𝐫𝐞 𝐚𝐟𝐭𝐞𝐫 𝐃𝐍𝐀 𝐫𝐞𝐩𝐥𝐢𝐜𝐚𝐭𝐢𝐨𝐧? Our new study “Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability” is out in 𝘚𝘤𝘪𝘦𝘯𝘤𝘦 𝘈𝘥𝘷𝘢𝘯𝘤𝘦𝘴. Led by @lleonie.bsky.social @biranalva.bsky.social 🧵 More below👇
19.02.2025 19:35 — 👍 112 🔁 53 💬 5 📌 5We need a genbank database
31.01.2025 11:48 — 👍 0 🔁 0 💬 0 📌 0Yeah I don’t really care how they made the plasmids. Just give me the genbank file.
30.01.2025 18:20 — 👍 22 🔁 4 💬 3 📌 0
Not only does Bennu contain all 5 of the nucleobases that form DNA and RNA on Earth and 14 of the 20 amino acids found in known proteins, the asteroid’s amino acids hold a surprise
https://go.nature.com/4hDtGhL
We hope that people find these ideas interesting, and that they maybe even provide new hypotheses to test experimentally. Thanks to Omer Gilan and @faulknerlab.bsky.social for help exploring these ideas over the last 5-6 years.
30.12.2024 22:58 — 👍 4 🔁 0 💬 0 📌 0One of the main goals of this review was to help clarify some of the confusion in the field about "epigenetics", specifically the interplay between TFs and histone mods. Here, we attempt to unify the role of transcription factors and histone modifications under a single conceptual framework.
30.12.2024 22:58 — 👍 2 🔁 0 💬 1 📌 0Chromatin memory has been appreciated for a long time, but we believe its implications have not been fully explored. The capacity for local, chromatin memory to stabilise non-encoded cell states has major implications for the evolution of cell types and formation of resistant cell states in cancer.
30.12.2024 22:58 — 👍 2 🔁 0 💬 2 📌 0
"Everything in epigenetics is circular". Here, we explore how memory that is stored locally on chromatin has a crucial role in stabilising cellular state, updating our view of the epigenetic landscape from one shaped by the genotype, to one moulded by experience
doi.org/10.1186/s130...

Collage of the 24 Development cover images in 2024
📢Vote for the 2024 Development cover image of the year
@dev-journal.bsky.social featured 24 cover images in 2024. Now is the chance to pick your favourite!
Browse through the images and vote by 13 Jan:
https://thenode.biologists.com/vote-for-the-2024-development-cover-image-of-the-year/
#DevBio
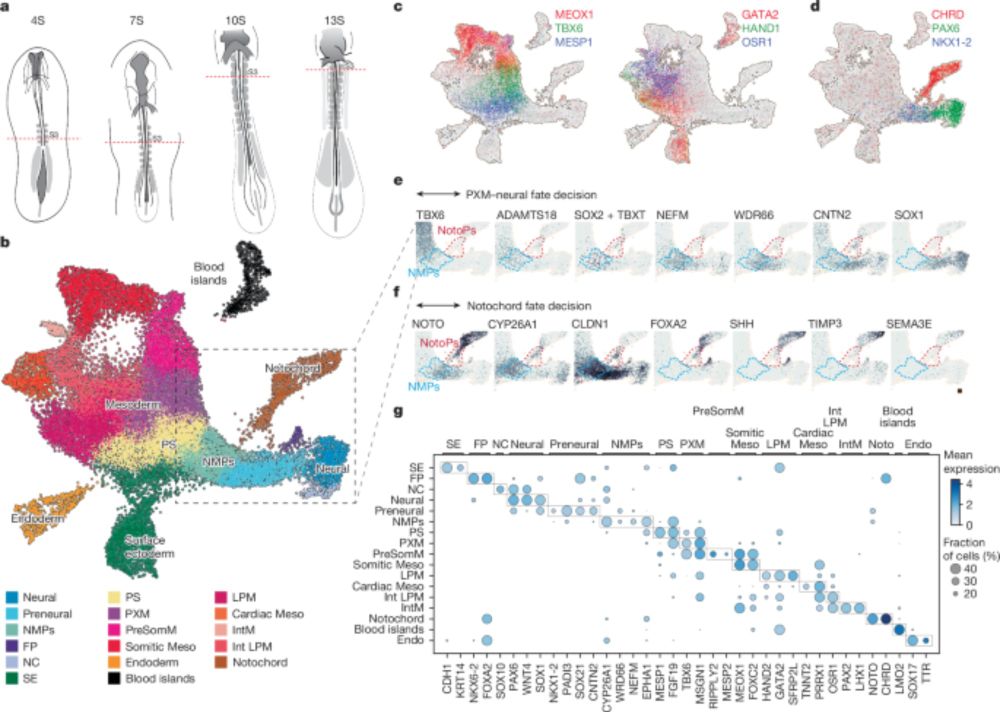
Our latest: how to generate the notochord
"Timely TGFβ signalling inhibition induces notochord"
A thread:
www.nature.com/articles/s41...

Crick researchers have recreated a tissue in the lab that acts like a developing embryo’s navigation system, directing cells where to build the spine and nervous system.
The findings were led by Tiago Rito with senior author @jamesbriscoe@bsky.social
www.crick.ac.uk/news/2024-12...

New in J R Soc Interface: How temporal dynamics of morphogen signalling increases positional precision in vertebrate neural tube development
Combining Shh & BMP signals over time provides significantly more spatial information than static gradients alone
doi.org/10.1098/rsif... #DevBio

Excited to share that my main PhD project has been published! 🎉 We systematically designed and optimized reporters for 86 transcription factors in parallel. If you're interested in using these optimized reporters for your own research, don’t hesitate to reach out!
13.12.2024 08:49 — 👍 20 🔁 8 💬 0 📌 0
What is the origin of universality in complex systems? This 1979 paper by Ken Wilson in @sciam.bsky.social
explains the Renormalization Group, one of the most powerful ideas in physics. Used to study everything from magnets and liquids to ecosystems and societies.
jstor.org/stable/24965...