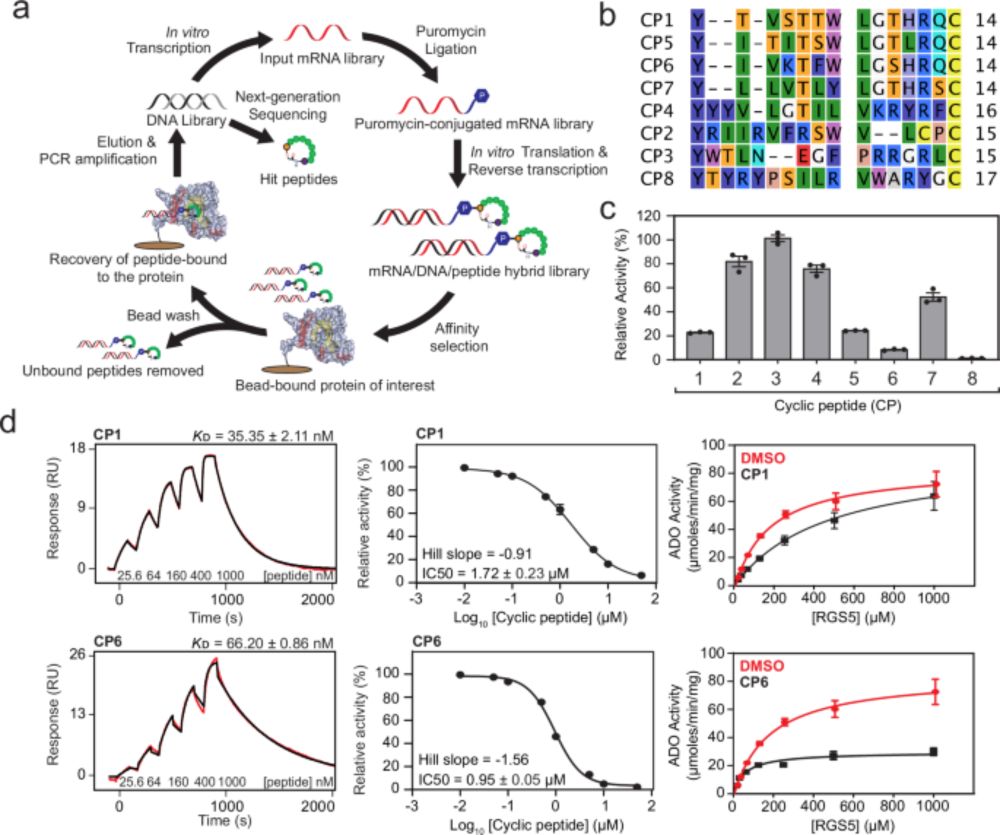
Developing leaves form a spatiotemporal oxygen gradient that is sensed through the oxygen sensing machinery and directs morphogenesis | New preprint from @daanweits.bsky.social and @syno2xis.bsky.social labs #N-degron #Cysteine #hypoxia @ispt-proteinterm.bsky.social www.biorxiv.org/content/10.1...
23.09.2025 07:38 — 👍 5 🔁 3 💬 1 📌 0
Methods in Enzymology | Volume 719: Protein Termini - Part B | ScienceDirect.com by Elsevier
Read the latest chapters of Methods in Enzymology at ScienceDirect.com, Elsevier’s leading platform of peer-reviewed scholarly literature
Two new volumes of Methods in Enzymology with 29 chapters/papers on Protein termini methods and reviews are now out! Thx to all authors!🙌 @ispt-proteinterm.bsky.social #N-degron #prenylation #myristoylation #formylation #arginylation #oxidation #acetylation www.sciencedirect.com/bookseries/m...
25.09.2025 09:17 — 👍 6 🔁 3 💬 1 📌 0

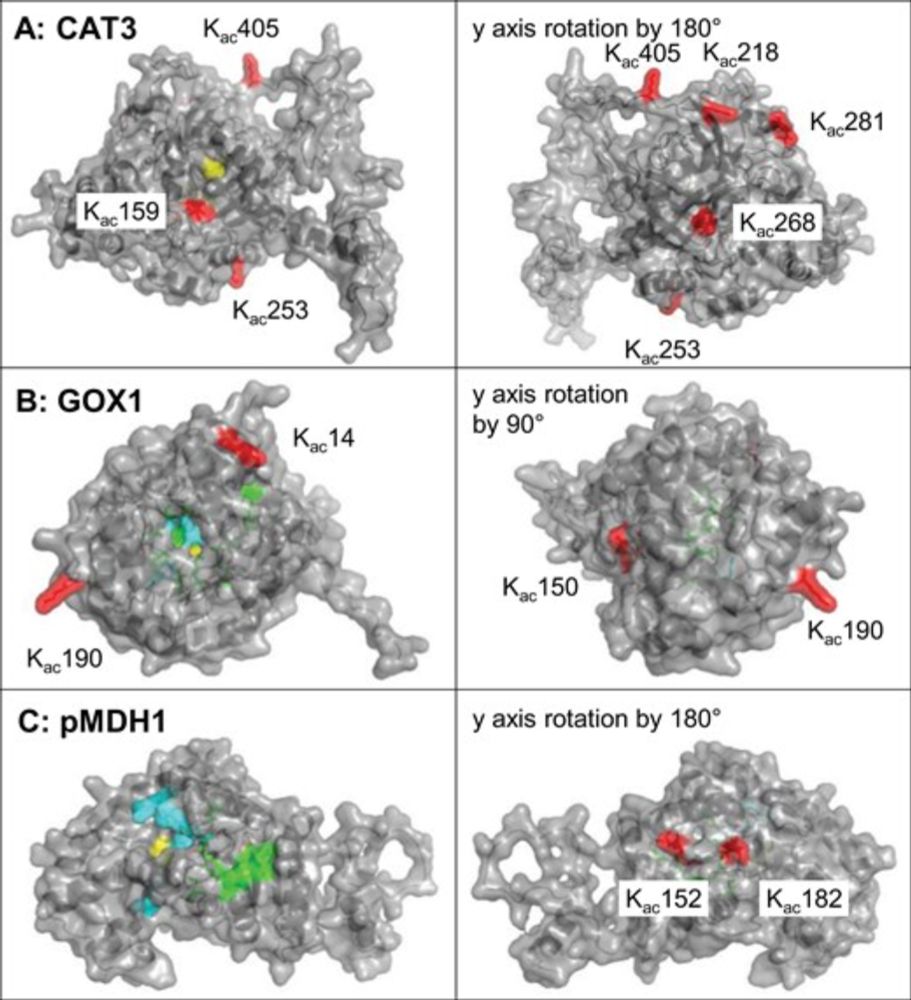
Protein N-terminal acetylation regulates the growth of rice plants | #NAA30 #NatC #rice #acetylation www.sciencedirect.com/science/arti...
25.09.2025 09:45 — 👍 3 🔁 3 💬 0 📌 0
Happy to share that our multi-omic analysis comparing two Arabidopsis accessions is now published 🌿 The first story from our @ukri.org BBSRC sLoLa #plant #proteostasis project. A great collaboration between @rothamsted.bsky.social @richardmott.bsky.social @ucl.ac.uk and Kathryn Lilley at Cambridge.
30.09.2025 09:56 — 👍 14 🔁 8 💬 0 📌 0
Daniel Gibbs Lab
Welcome to the lab website of Professor Daniel Gibbs
@ University of Birmingham
We are advertising a PhD project investigating a cellular mechanism that ensures protein quality control during mRNA translation in plants. Please get in touch on here or via email if you are interested and I can provide further details!
Lab website: sites.google.com/site/danielg...
14.10.2025 14:07 — 👍 32 🔁 36 💬 0 📌 0
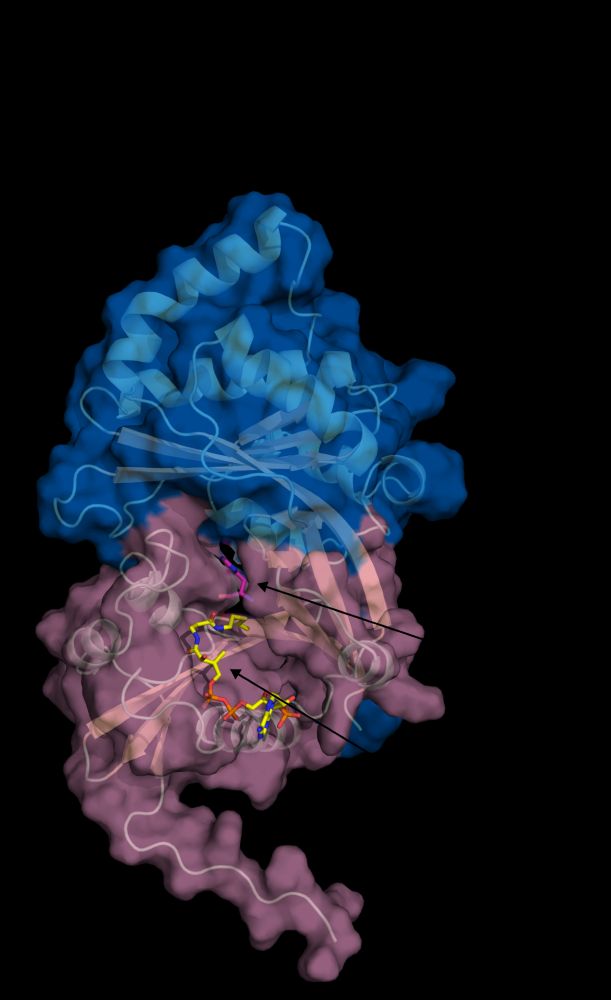
Acetylhistidine in our blood can prevent kidney disease and is produced by the HisAT (NAT16) enzyme displaying a rare double-GNAT structure. Great efforts by Matti Myllykoski and the rest of the team at @unibergen.bsky.social to get this published. #acetyltransferase www.nature.com/articles/s41...
01.07.2025 13:44 — 👍 9 🔁 7 💬 0 📌 0

Two heroes of the COST ProteoCure network: Oliver Coux and Laetitia Poidevin. Many thanks from all of us! #ProteoCure2025 @costprogramme.bsky.social
26.05.2025 10:29 — 👍 3 🔁 2 💬 0 📌 0
'
A postdoc position is available in our lab to study ADP-ribosylation and ubiquitylation signalling! We are looking for someone with experience in structural biology and biochemistry. The application deadline is June 20th.
my.corehr.com/pls/uoxrecru...
24.05.2025 05:29 — 👍 20 🔁 23 💬 1 📌 2

Congrats to Corentin Bouvier, awardee of the FEBS Journal prize at the annual COST ProteoCure meeting. @costprogramme.bsky.social @febsj.bsky.social #ProteoCure2025
24.05.2025 08:22 — 👍 8 🔁 3 💬 0 📌 0

Congrats to Mónica Pozo Rodriguez, awardee of the FEBS Open Bio Poster prize at the annual COST ProteoCure meeting. @costprogramme.bsky.social @febspress.bsky.social #ProteoCure2025
24.05.2025 08:23 — 👍 6 🔁 3 💬 0 📌 0

Congrats to Simon Tack, awardee of the FEBS Journal Oral Presentation prize at the annual COST ProteoCure meeting.
@costprogramme.bsky.social @febsj.bsky.social
#ProteoCure2025
24.05.2025 08:25 — 👍 8 🔁 3 💬 0 📌 0

Opening of the 2025 ProteoCure #COST meeting on Crete by Rosa Farras. Looking forward to an exciting meeting.
21.05.2025 07:53 — 👍 2 🔁 2 💬 0 📌 0
Nobel Laureate Aaron Ciechanover gave a historic overview on the discovery of ubiquitylation and finished with some very recent data from his lab at the annual #ProteoCure meeting.
21.05.2025 07:57 — 👍 5 🔁 1 💬 0 📌 0
Michael Glickman presented his latest data on one of our favorite proteins UBB+1 and its role in Alzheimer's disease at the annual meeting of COST ProteoCure in Heraklion.
21.05.2025 07:59 — 👍 4 🔁 2 💬 0 📌 0
Starting the morning at #COST #ProteoCure2025 annual meeting with Elke Krüger talking about Proteasomopathies. @costprogramme.bsky.social
22.05.2025 06:29 — 👍 2 🔁 1 💬 0 📌 0
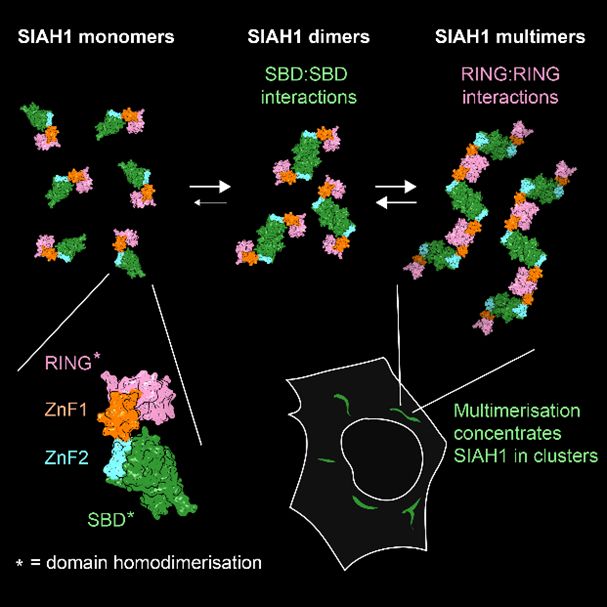
Happy to share our new study showing that the Ub E3 ligase SIAH1, known to dimerise via its C-term SBD, also dimerises via its N-term RING. When these tendencies combine, full-length SIAH1 forms multimers, which might explain its clustering in cells & preference for aggregated/multimeric substrates.
06.02.2025 09:29 — 👍 56 🔁 16 💬 4 📌 1

Genomic information pathways and their numerical linkages in relation to the Central Dogma. The boxes show different omic levels in Col-0 and Can-0. Black arrows indicate the flow of information implied by the Central Dogma starting from CDS (peach) via gbM (green) to mRNA (light blue), ribosome-associated mRNA (yellow) and finally protein (pink). The strength of Pearson correlation between levels is shown both by the numbers and by the thickness of the corresponding black arrows, and represents the correlation between quantitative expression levels (except in the case of CDS where is represents the correlation derived from the multiple regression codon frequency model, i.e. the square root of the fraction of variance explained). The merged arrows connecting CDS, gbM and mRNA to protein show the result of combining information into a single model; note that their combined correlation of 0.720 is less than the sum of their individual effects. The correlation of tRNA levels (lilac) with genome-wide codon and amino acid frequencies is shown on the left. The correlations between corresponding Col-0 and Can-0 omic levels are shown next to the grey double-headed arrows.
Happy to start 2025 by sharing our new preprint biorxiv.org/cgi/content/... -the first paper from our BBSRC collaborative project with
@richardmott.bsky.social and Kathryn Lilley to understand what controls protein abundance in plants. 🧵 1/9
13.01.2025 15:42 — 👍 44 🔁 15 💬 4 📌 1
New insight into the plant #N-degronpathway from its co-discoverer, Andreas Bachmair and colleagues. The Arabidopsis N-recognin PRT6 has broader specificity than previously thought.
13.01.2025 15:50 — 👍 8 🔁 4 💬 0 📌 0
Congratulations!
15.12.2024 15:22 — 👍 1 🔁 0 💬 0 📌 0

Job Opportunity: Assistant Engineer in N-terminomics!Join our team at I2BC, Université Paris-Saclay (20 km south of Paris)! Looking for a motivated person with expertise in mass spectrometry (N-terminal proteomics, quantitative analysis). Please share! @proteintermini
#proteomics.
03.12.2024 17:29 — 👍 0 🔁 1 💬 0 📌 0
Since 1911, we have supported the molecular bioscience community, promoting the sharing of knowledge and enhancing career development.
Discover more about the Society: https://linktr.ee/biochemicalsociety
Assistant Prof at UWaterloo in Biology. Study autophagy & protein lipidation (S-acylation & N-myristoylation) in ALS & Huntington disease. Expect science, food, some cdnpoli, & dog pics
https://neurdyphagylab.squarespace.com/
A sound proteome for a sound body: targeting proteolysis for proteome remodeling
Senior Editor at Molecular Cell
Non-profit society of scientists devoted to understanding the molecular mechanisms and biological role of protein termini in plants, animals, fungi and bacteria. Conference every second year. #acetylation #arginylation #oxidation #N-degron #C-degron
University of Edinburgh research group studying how protein modifications contribute to plant resilience in a changing environment.
Structural biologist - currently ubiquitin ligases, previously DNA replication and bacterial metabolism
Research Associate - Clausen Lab, IMP Vienna
Senior lecturer & Researcher in #microbiology, #cellbiology #platelets at Sheffield Hallam University, Sheffield, UK.
Speaks English, Français, Mauritian Creole (from the land of the dodos) & basic Hindi. She/her #WomenInStem #EDI
Professor of Biochemistry at St. Louis University School of Medicine. Passionate about mechanistic & structural enzymology, electron transfer & nucleic acid binding enzymes. Fascinated by Cryo-EM, kinetics, single molecules and student training. Golf Nut!
Interested in tech, politics, design
Junior Project Leader, Ramón y Cajal Fellow. Plants, pathogens and proteostasis at CNB-CSIC (Madrid)
Non-for-profit publication of research in molecular, cell & organismal biology - as part of @embopress.org
Fast-fair-transparent / Open Access / Open Science
BioGRID is a comprehensive #biomedical repository for curated #protein, #genetic and #chemical interactions | Funding @NIH_ORIP
& @CIHR_IRSC | #CRISPR #opendata
https://thebiogrid.org
https://orcs.thebiogrid.org
Professor and Director, Dept. of Biophysics & Biophysical Chemistry
Johns Hopkins School of Medicine, structural biologist, lover of chromatin and ubiquitin, fan of the active voice.
Professor of Proteomics. Currently at Newcastle University UK. he/him
Here in personal capacity.
Scientist interested in ubiquitin and ubiquitin-like proteins in host defense to intracellular pathogens; based at National Jewish Health in Denver. Excited about proteomics, Listeria, Francisella and SARS-CoV-2. Views are my own
Structural biologist and biochemist, interested in ubiquitin enzymes and DNA repair & regulation
Associate Professor at Oregon Health & Science University studying microbial interference in host ubiquitin signaling. Current ASM Northwest Branch President. Formerly @UWBiochemistry and @MRC_LMB.
VP & Head of Induced Proximity Platform, @Amgen | Scientist | Father