Dr. Margaret Oakley Dayhoff
I took biochem in 2001, and for nearly 20 years read amino acid sequences daily… and I never knew Dayhoff named them or even the logic behind things like Q until last Friday (h/t Mike Janech). Also, this is another big Dayhoff moment for me. She was incredible!
#proteomics #bioinformatics
24.11.2024 12:39 — 👍 198 🔁 79 💬 14 📌 7
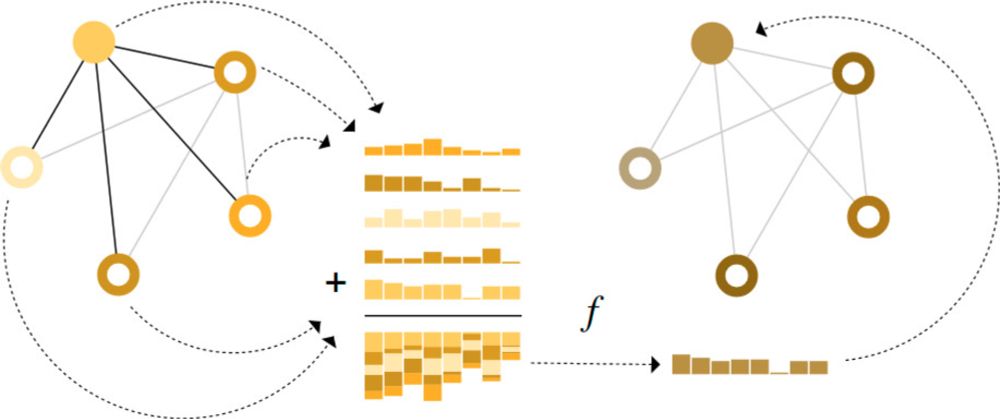
A Gentle Introduction to Graph Neural Networks
What components are needed for building learning algorithms that leverage the structure and properties of graphs?
Always so impressed by how good this intro to graph neural nets is. They did such a good job of broadly covering the field without diving into a million papers. I love that they build intuition for how designing GNN architectures is tricky, wish more ML posts did that.
20.11.2024 16:42 — 👍 14 🔁 0 💬 0 📌 0
You could run nvidia-smi in the terminal while your model is running to see if your vram is full.
20.11.2024 13:50 — 👍 7 🔁 0 💬 0 📌 0
Ah I wasn't saying that your pcie bandwidth would be abnormally low but rather that you might not be able to fit everything into the rtx4090s vram and so you'd have to make a lot of transfers between cpu ram & vram which is slow and would leave the GPU waiting for data most of the time. :)
20.11.2024 13:22 — 👍 1 🔁 0 💬 1 📌 0
This feels like it's just due to the rtx4090 being pcie bandwidth limited for some reason. The peak fp16 compute for an m4 max should be 34 tflops while the rtx is 82 tflops and the vram bandwidth is twice as fast in an rtx.
What happens if you run the model with a smaller resolution img or fp8?
20.11.2024 12:47 — 👍 1 🔁 0 💬 1 📌 0
Not restricting it to the fully de novo case, even an example where a model makes a few mutations in a wild-type sequence is fine.
Totally agree that all the work showing some activity in de novo sequences is super impressive.
20.11.2024 10:16 — 👍 0 🔁 0 💬 0 📌 0
Intentionally didn't want it to be too high a bar. A single substitution is totally fine as long as the model isn't constrained to mutate a few clearly impactful residues in the active site for example.
20.11.2024 09:58 — 👍 0 🔁 0 💬 0 📌 0
I haven't been able to find a paper that:
1. uses ML to propose enzyme sequence
2. measures kcat of designed enzyme and highly similar sequence in the train set
3. shows that the designed enzyme is faster than all other enzymes that catalyze the same reaction in the train set with a known rate
20.11.2024 09:53 — 👍 0 🔁 0 💬 1 📌 0
Can't be ruled out 100%, but some designed sequences are going to be more plausibly out-of-distribution than others.
20.11.2024 09:33 — 👍 0 🔁 0 💬 1 📌 0
Depends on how big your dataset is. I'm fine with a training set that includes all naturally occuring sequences where most don't have associated kcats for example. I just want to avoid cases where you can just pick a wild-type protein from the same family to get an improved enzyme.
19.11.2024 23:16 — 👍 0 🔁 0 💬 1 📌 0
Has a machine learning model ever successfully designed an enzyme that's 5x faster than the sequences in its training set?
Specifically looking for an experimentally verified example where the model is the decision maker rather than a human assistant.
19.11.2024 19:01 — 👍 2 🔁 0 💬 2 📌 0
Would love to be added to this. :)
19.11.2024 10:18 — 👍 1 🔁 0 💬 1 📌 0
I'd love to be added. I create microfluidic devices and large-scale datasets which I use to train protein language models.
19.11.2024 10:16 — 👍 1 🔁 0 💬 1 📌 0
PhD student Winter lab @ AITHYRA/ CeMM Vienna | interested in using structural and synthetic biology to engineer cellular decision making | previously Baker lab @ IPD and Taylor lab @ mpiib-berlin
ELLIS PhD, University of Tübingen | Data-centric Vision and Language @bethgelab.bsky.social
Website: adhirajghosh.github.io
Twitter: https://x.com/adhiraj_ghosh98
Zon lab NCI K00 postdoc | pediatric sarcoma PhD | Luger lab chromatin biochem MS | transcription and melanoma in #zebrafish 🐠 | she/her
Reverse engineering neural networks at Anthropic. Previously Distill, OpenAI, Google Brain.Personal account.
assistant prof at USC Data Sciences and Operations and Computer Science; phd Cornell ORIE.
data-driven decision-making, operations research/management, causal inference, algorithmic fairness/equity
bureaucratic justice warrior
angelamzhou.github.io
CTO & Co-Founder at Coefficient Bio. Ex-Prescient Design • Genentech. Advisor to Atomscale & Guide Labs
ncfrey.github.io | ncfrey.substack.com
Associate Prof. at WashU School of Medicine; biophysics/biochem/evolution of intrinsically disordered proteins. How does nature encode function without a stable structure? We work in vivo / in vitro / in silico. He/him.
https://www.holehouselab.com/
Associate Professor (UHD) at the University of Amsterdam. Probabilistic methods, deep learning, and their applications in science in engineering.
PI at the Uni Medical Center Göttingen (UMG), Founder and CTO of NanoTag Biotechnologies. Using nanobodies for super-resolution microscopy in neurosciences. Opinions are my own.
Postdoctoral researcher @ParisBrainInst / @InstitutCerveau working on oligodendroglia and CNS myelination. Cell biology teacher @supbiotech. Biotech engineer.
Neuroimmunology lab at UC Irvine studying the role of microglia in Alzheimer’s disease and developing iPSC-microglia based therapeutic approaches.
Ph.D. Candidate studying how Group A Streptococcus adapts to heme stress during invasive infections
Bacterial genetics | Bacterial metabolism | AMR | SynBio
He/him
Views are my own
Assistant Professor in Soft Matter Physics at Aalto University, Finland | Forces and flow in mesoscale living, fluid, & soft matter | ERC StG laureate & Research Council of Finland Fellow | EPL co-editor
aalto.fi/living-matter
Research Scientist @ Google DeepMind. Physics of learning, ML / AI, condensed matter. Prev Ph.D. Physics @ UC Berkeley.
Cofounder/CEO Octant
BoD Ginkgo Bioworks
Defense Science Board for Emerging Biotech
Fmr: Associate Professor, UCLA
Neuroimmune axis, mucosal immunology, microbial ecology and evolution, geobiology.
Anti-fascist. Apparently I live in a country where that has to be explicitly stated.
Father and scientist. Doing dad stuff in Brooklyn and science at New York Genome Center.
𝘈𝘴𝘴𝘰𝘤𝘪𝘢𝘵𝘦 𝘋𝘪𝘳𝘦𝘤𝘵𝘰𝘳, 𝘚𝘱𝘢𝘵𝘪𝘢𝘭 𝘎𝘦𝘯𝘰𝘮𝘪𝘤𝘴
𝘈𝘳𝘤𝘩𝘪𝘷𝘦 𝘰𝘧 𝘮𝘺 𝘵𝘪𝘮𝘦 𝘢𝘵 𝘵𝘩𝘦 𝘰𝘭𝘥 𝘱𝘭𝘢𝘤𝘦 @𝘮𝘢𝘯𝘪𝘮𝘰𝘵𝘢𝘴𝘰𝘭𝘥𝘴𝘵𝘶𝘧𝘧.𝘣𝘴𝘬𝘺.𝘴𝘰𝘤𝘪𝘢𝘭
@𝘮𝘢𝘯𝘪𝘮𝘰𝘵𝘢 𝘰𝘯 𝘮𝘰𝘴𝘵 𝘱𝘭𝘢𝘵𝘧𝘰𝘳𝘮𝘴
Principal Researcher in BioML at Microsoft Research.
Professor of Biotechnology at Aston Uni. Interested in all things membranes and biotech. All opinions my own (not sure anyone else would want them).
