PUCHIK means 'balloon' in Armenian, which is home for Hrach and is also a well-suited name for the nanoscale objects that we hope to facilitate the understanding of with this code. 10/10
13.02.2025 08:55 — 👍 0 🔁 0 💬 0 📌 0@lorenzlab1.bsky.social
@lorenzlab1.bsky.social
PUCHIK means 'balloon' in Armenian, which is home for Hrach and is also a well-suited name for the nanoscale objects that we hope to facilitate the understanding of with this code. 10/10
13.02.2025 08:55 — 👍 0 🔁 0 💬 0 📌 0The efforts of so many people have resulted in numerous manuscripts being published in close succession. Hrach Ishkhanyan & Alejandro Santana Bonilla did all of the hard work in this case. 9/10
13.02.2025 08:55 — 👍 0 🔁 0 💬 1 📌 0If you wish to listen to a podcast summarising our manuscript, then here is a link: bit.ly/3QcUY2F. 8/10
13.02.2025 08:55 — 👍 0 🔁 0 💬 1 📌 0
PUCHIK also assists in linking MD simulation data to experimental data by providing accurate definitions of nanoparticle shapes, which are needed for modelling neutron scattering experiments. The code is freely available here: bit.ly/4hYh7Oi. 7/10
13.02.2025 08:55 — 👍 0 🔁 0 💬 1 📌 0PUCHIK is built for computational efficiency using SciPy, MDAnalysis, and Cython, and is designed to be user-friendly and modular. 6/10
13.02.2025 08:55 — 👍 0 🔁 0 💬 1 📌 0PUCHIK can be used to analyse each aggregate separately in simulated systems with multiple nanoparticles. In particle, this software provides accurate descriptions of aspherical nanoparticles, which are poorly characterised by methods employed in our PySoftK software. 5/10
13.02.2025 08:55 — 👍 0 🔁 0 💬 1 📌 0By accurately accounting for the location of the interface of the aggregate, one can also accurately determine the amount of molecular cargo solubilised within the nanoparticles (e.g. small molecule or peptide therapeutic or any other desirable molecular cargo). 4/10
13.02.2025 08:55 — 👍 0 🔁 0 💬 1 📌 0The software enables the calculation of key properties, such as densities of (and therefore the internal structure) the molecules which are found within the nanoparticle and in its local environment, nanoparticle volume and surface area. 3/10
13.02.2025 08:55 — 👍 0 🔁 0 💬 1 📌 0PUCHIK employs both convex hulls and alpha shapes to define the nanoparticle interface, allowing for a detailed representation of solid, hollow, and mesoporous nanoparticles. This is crucial as surface effects are dominant in nanoparticle functionality. 2/10
13.02.2025 08:55 — 👍 0 🔁 0 💬 1 📌 0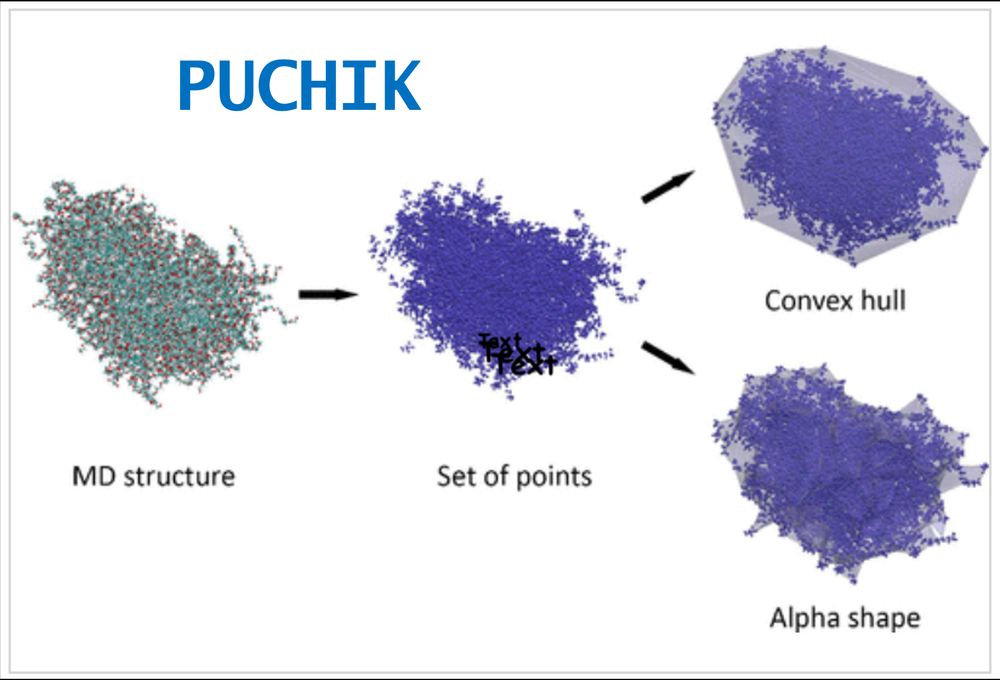
Our latest manuscript to be published introduces PUCHIK (bit.ly/3QeqbT4), a novel & efficient Python package designed for the analysis of molecular dynamics simulations of nanoparticles, w/ a focus on accurately describing the interfaces of aspherical nanoparticles. 1/10
13.02.2025 08:55 — 👍 1 🔁 1 💬 1 📌 0Thanks to the amazing Alejandro Santana-Bonilla , Rob Ziolek & Raquel López-Ríos de Castro for doing all of the hard work! 7/7
11.02.2025 23:54 — 👍 0 🔁 0 💬 0 📌 0The code and tutorials can be found via the PySoftK website: bit.ly/3CXXML3 ... If you want to here a podcast summarising the latest in PySoftK then here is a link: bit.ly/3WUJSmT 6/7
11.02.2025 23:54 — 👍 0 🔁 0 💬 1 📌 0PySoftK v1.0 provides an automated approach to investigate interfaces in soft matter systems, interactions that govern structure within soft matter, and soft matter self-assembly, and will help to accelerate the computer-aided design of new materials. 5/7
11.02.2025 23:54 — 👍 0 🔁 0 💬 1 📌 0Finally, it has some basic tools for calculating the radius of gyration and eccentricity of self-assembled structures. 4/7
11.02.2025 23:54 — 👍 0 🔁 0 💬 1 📌 0Also there is a tool that correctly accounts for periodic boundary conditions when analysing self-assembled structures, even if these structures are larger than 1/2 of the simulation box (this is not possible with any other tool & allows accurate analyses of these structures) 3/7
11.02.2025 23:54 — 👍 0 🔁 0 💬 1 📌 0The software now includes tools that can quantify the self-assembly of molecules, analyse the interfaces of nanoparticles (spherical & sphere-like), investigate molecular interactions like ring stacking, solvation and intermolecular contacts. 2/7
11.02.2025 23:54 — 👍 0 🔁 0 💬 1 📌 0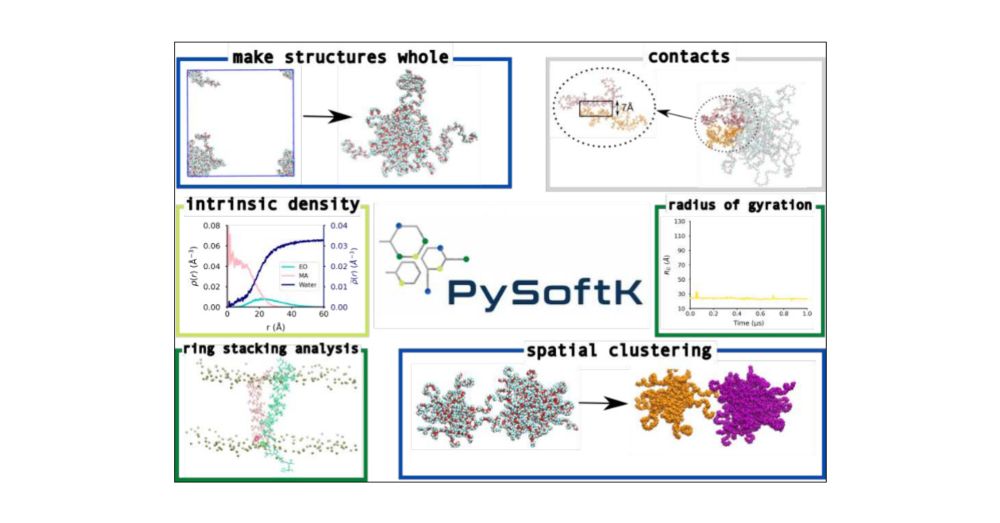
Our latest manuscript presents the most recent tools available in the PySoftK software package (bit.ly/4jQps8i). PySoftK now helps scientists analyse molecular simulations of soft materials, w/ tools for analysing interfaces, molecular interactions, & self-assembly 1/7
11.02.2025 23:54 — 👍 1 🔁 1 💬 1 📌 0This is another nice manuscript which has resulted from my continued collaboration with Max Ryadnov (NPL) & Bart Hoogenboom (@ucl.ac.uk). If you are interested in listening to a podcast summarising the paper then here you go: bit.ly/41aueGv 3/3
10.02.2025 19:52 — 👍 0 🔁 0 💬 0 📌 0This process, which we observed using advanced imaging and molecular dynamics simulations, is similar to how some other antimicrobial agents work and offers a potential new approach to fighting drug-resistant bacteria. 2/3
10.02.2025 19:52 — 👍 0 🔁 0 💬 1 📌 0
Our latest manuscript has been published in
Langmuir: bit.ly/4hMNBuy. In this manuscript we show that thanatin, a naturally occurring antimicrobial peptide, punches holes & disrupts bacterial membranes through multiple mechanisms, causing them to break apart 1/3
@kingsnmes.bsky.social @kingslsm.bsky.social
29.01.2025 19:38 — 👍 0 🔁 0 💬 0 📌 0I generated a NotebookLM podcast summarising the paper, to listen - here is a link: bit.ly/3PWshqy. This was a great collaboration w/ Anabella Gaspar & Megan Bester from Pretoria & @profjamesmason.bsky.social w/ Rosalind Van Wyk doing all the hard work & Miruna Serian supporting on the MD.
29.01.2025 19:38 — 👍 0 🔁 0 💬 1 📌 0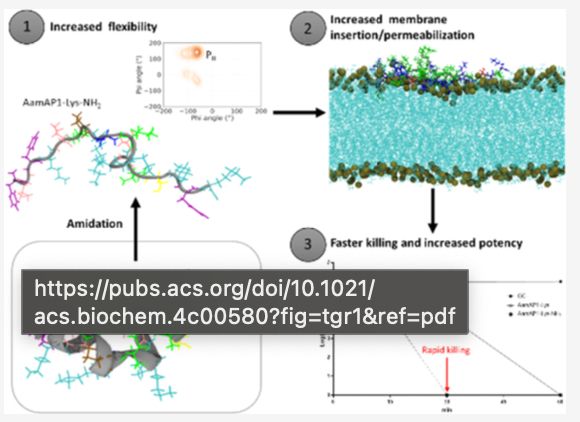
Our latest manuscript is now available in Biochemistry @acs.org (bit.ly/3Ct1PSm) In it we show that C-terminal amidation of an antimicrobial peptide increases its conformational flexibility, which improves membrane penetration, leads to faster bacterial killing & better selectivity for pathogens.
29.01.2025 19:38 — 👍 8 🔁 4 💬 1 📌 0