Figure 1 - Live imaging of endogenously tagged YAP-miRFP670, mScarlet-I-SOX2 and CDX2-eGFP during the first fate decision in preimplantation embryos. (A) A developmental timeline of trophectoderm (TE) and inner cell mass (ICM) specification. The first cell differentiation event, between the TE (green) and ICM (purple), initiates at the 16-cell stage, establishing distinct inner, ICM, and outer, TE, cell populations in the 32-cell stage embryo. Embryos in this study are imaged until the 32-cell stage. Days post-fertilization are denoted by embryonic day. (B) Sox2 and Cdx2 expression is regulated by YAP. In ICM cells, YAP is phosphorylated and retained in the cytoplasm, facilitating Sox2 expression. In TE cells, YAP is predominantly localized to the nucleus where it activates expression of Cdx2. (C) Time-lapse images of a representative embryo expressing YAP-miRFP670, mScarlet-I-SOX2 and H2B-miRFP670 from the 8-cell through the late 32-cell stage. A z-stack was acquired every 15 min for YAP and H2B channels, and every 30 min for the SOX2 channel. Maximum intensity projections (MIPs) are shown. Unit of time is hours. Scale bar: 10 µm. (D) Time-lapse images of a representative embryo expressing YAP-miRFP670, CDX2-eGFP and H2B-miRFP670 from the 8-cell stage through the late 32-cell stage. Scale bar: 10 µm.
Mathematical modelling of the first fate decision
This Research Highlight showcases the work from Maria Avdeeva, Madeleine Chalifoux, Bradley Joyce, Stanislav Y. Shvartsman, Eszter Posfai @eposfai.bsky.social :
journals.biologists.com/dev/article/...
10.09.2025 15:42 — 👍 15 🔁 4 💬 1 📌 0
The #devbio project that encompassed the majority of my postdoc is finally out! Huge thanks to my coauthors and our collaborators without whom this work would not have been possible, and my adviser Eszter Posfai who let me play with a light sheet microscope and some colorful mice :)
03.08.2025 16:45 — 👍 7 🔁 2 💬 0 📌 0
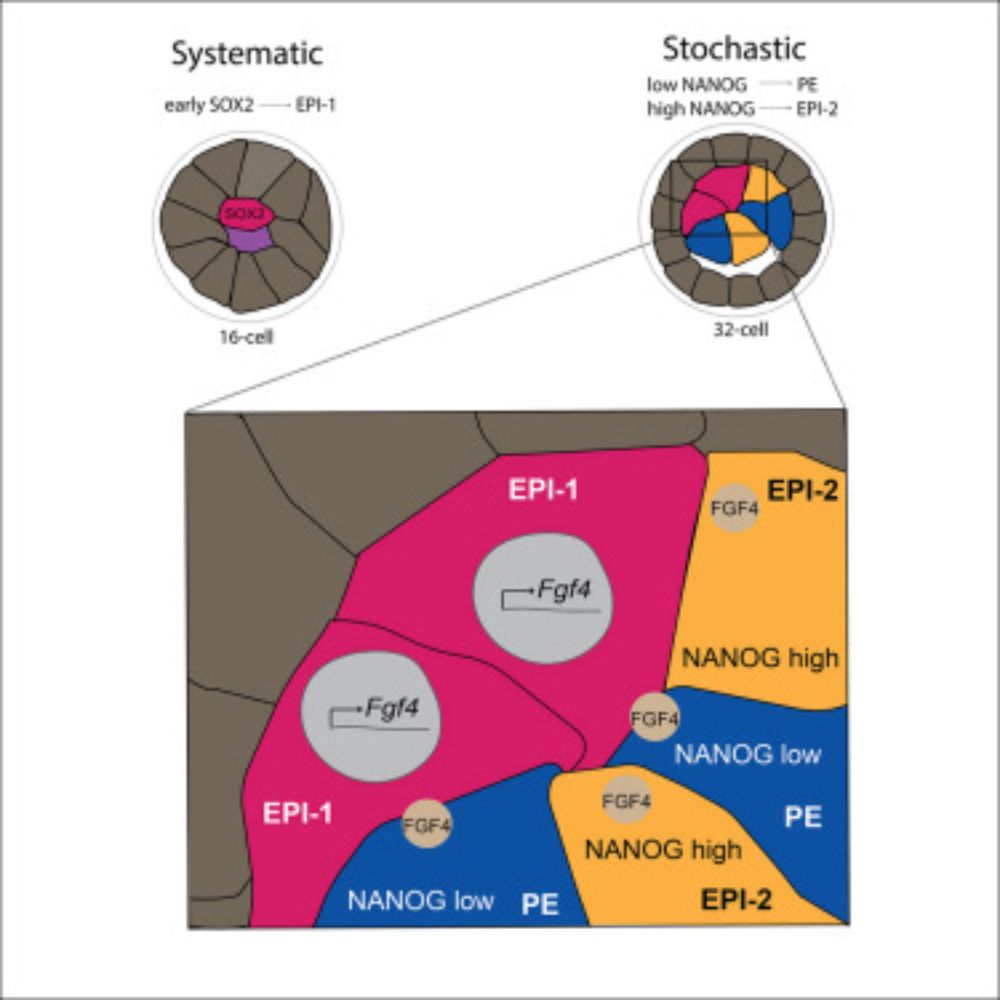
Whether the trajectory is maintained towards PE or switches to Epi is in part predicted by the level of another TF, NANOG, which in turn seems to behave in a stochastic manner.
02.08.2025 19:31 — 👍 3 🔁 0 💬 1 📌 0
We then showed that FGF signaling, with the ligand likely from the primary Epi lineage, sets all other cells on the course towards PE differentiation. This trajectory however switches in some lineages, within a defined developmental window, giving rise to a second wave of Epi cells.
02.08.2025 19:31 — 👍 2 🔁 0 💬 1 📌 0
We found the emergence of a founder, termed primary Epi cell lineage as the first symmetry breaking event. Moreover, this primary Epi cell was marked by early expression of SOX2, a TF linked to the segregation of the previous fate decision. Therefore, symmetry breaking is linked to lineage history.
02.08.2025 19:31 — 👍 2 🔁 0 💬 1 📌 0
We simultaneously live image endogenously tagged transcription factors (up to 3 of them!) key to this fate decision, which allowed us not only to analyze the lineage history of cells, but also to directly visualize how individual cells choose their fates in developing embryos.
02.08.2025 19:31 — 👍 2 🔁 0 💬 1 📌 0
Thrilled to share our work using live imaging to understand how Epiblast (future embryo proper) and Primitive Endoderm (future extraembryonic tissues) cell fates segregate in the preimplantation mouse embryo. Gargantuan effort led by amazing @rpkimyip.bsky.social, David Denberg and Denis Faerberg!
02.08.2025 19:31 — 👍 37 🔁 12 💬 1 📌 1
pYtags - our in vivo biosensors for receptor tyrosine kinases - in all their beauty on the cover of Cell Reports! Read more here: www.cell.com/cell-reports...
22.07.2025 20:52 — 👍 90 🔁 20 💬 5 📌 3
We’re hiring! Our lab at WashU has an opening for a Senior/Staff Scientist position. If you're passionate about ovarian physiology, oocyte quality, and excited to contribute to a dynamic and growing team, we’d love to hear from you! Reach out to me directly for more information.
21.07.2025 22:39 — 👍 3 🔁 7 💬 1 📌 0
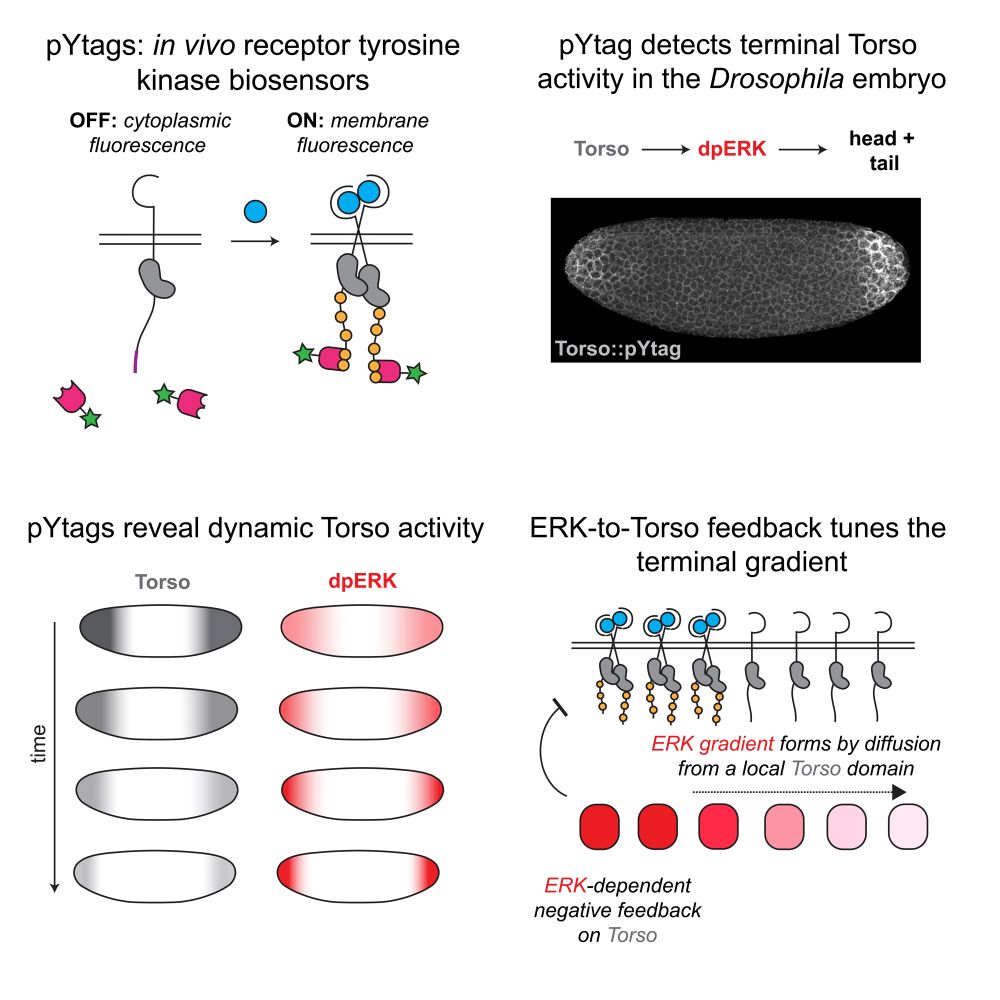
Excited to share that our work building in vivo biosensors for receptor tyrosine kinases (pYtags!) is now out www.cell.com/cell-reports...
01.07.2025 13:58 — 👍 40 🔁 20 💬 3 📌 0

Generative model for the first cell fate bifurcation in mammalian development
The first cell fate bifurcation in mammalian development directs cells toward either the trophectoderm (TE) or inner cell mass (ICM) compartments in preimplantation embryos. This decision is regulated by the subcellular localization of a transcriptional co-activator YAP and takes place over several progressively asyn-chronous cleavage divisions. As a result of this asynchrony and variable arrangement of blastomeres, reconstructing the dynamics of the TE/ICM cell specification from fixed embryos is extremely challenging. To address this, we developed a live imaging approach and applied it to measure pairwise dynamics of nuclear YAP and its direct target genes, CDX2 and SOX2, key transcription factors of TE and ICM, respectively. Using these datasets, we constructed a generative model of the first cell fate bifurcation, which reveals the time-dependent statistics of the TE and ICM cell allocation. In addition to making testable predictions for the joint dynamics of the full YAP/CDX2/SOX2 motif, the model revealed the stochastic nature of the induction timing of the key cell fate determinants and identified the features of YAP dynamics that are necessary or sufficient for this induction. Notably, temporal heterogeneity was particularly prominent for SOX2 expression among ICM cells. As heterogeneities within the ICM have been linked to the initiation of the second cell fate decision in the embryo, understanding the origins of this variability is of key significance. The presented approach reveals the dynamics of the first cell fate choice and lays the groundwork for dissecting the next cell fate bifurcations in mouse development. ### Competing Interest Statement The authors have declared no competing interest.
The second paper, incorporating a probabilistic modeling approach, investigates the relationship between YAP dynamics and two of its downstream transcription factor targets, SOX2 (ICM) and CDX2 (TE).
www.biorxiv.org/content/10.1...
05.03.2025 02:54 — 👍 3 🔁 0 💬 0 📌 0

Geometric, cell cycle and maternal-to-zygotic transition-associated YAP dynamics during preimplantation embryo development
During the first cell fate decision in mammalian embryos the inner cell mass cells, which will give rise to the embryo proper and other extraembryonic tissues, segregate from the trophectoderm cells, the precursors of the placenta. Cell fate segregation proceeds in a gradual manner encompassing two rounds of cell division, as well as cell positional and morphological changes. While it is known that the activity of the Hippo signaling pathway and the subcellular localization of its downstream effector YAP dictate lineage specific gene expression, the response of YAP to these dynamic cellular changes remains incompletely understood. Here we address these questions by quantitative live imaging of endogenously tagged YAP while simultaneously monitoring geometric cellular features and cell cycle progression throughout cell fate segregation. We apply a probabilistic model to our dynamic data, providing a quantitative characterization of the mutual effects of YAP and cellular relative exposed area, which has previously been shown to correlate with subcellular YAP localization in fixed samples. Additionally, we study how nuclear YAP levels are influenced by other factors, such as the decreasing pool of maternally provided YAP that is partitioned to daughter cells through cleavage divisions, cell cycle-associated nuclear volume changes, and a delay after divisions in adjusting YAP levels to new cell positions. Interestingly, we find that establishing low nuclear YAP levels required for the inner cell mass fate is largely achieved by passive cell cycle-associated mechanisms. Moreover, contrary to expectations, we find that mechanical perturbations that result in cell shape changes do not influence YAP localization in the embryo. Together our work identifies how various inputs are integrated over a dynamic developmental time course to shape the levels of a key molecular determinant of the first cell fate choice. ### Competing Interest Statement The authors have declared no competing interest.
The first paper looks at how different inputs, such as cell position and shape, cell cycle progression and transitioning from maternal to zygotic resources during early development impact the nuclear levels of YAP, a key regulator this fate decision.
www.biorxiv.org/content/10.1...
05.03.2025 02:54 — 👍 2 🔁 0 💬 1 📌 0
A duo of preprints on the dynamics of the first cell fate decision in mouse by Madeleine Chalifoux (first grad student in the lab!) and Maria Avdeeva (Flatiron).
We use quantitative live imaging of key cell fate determinants to follow the segregation of inner cell mass and trophectoderm lineages.
05.03.2025 02:54 — 👍 25 🔁 11 💬 1 📌 0
Congrats, looking fwd to reading!!
12.02.2025 00:18 — 👍 1 🔁 0 💬 1 📌 0
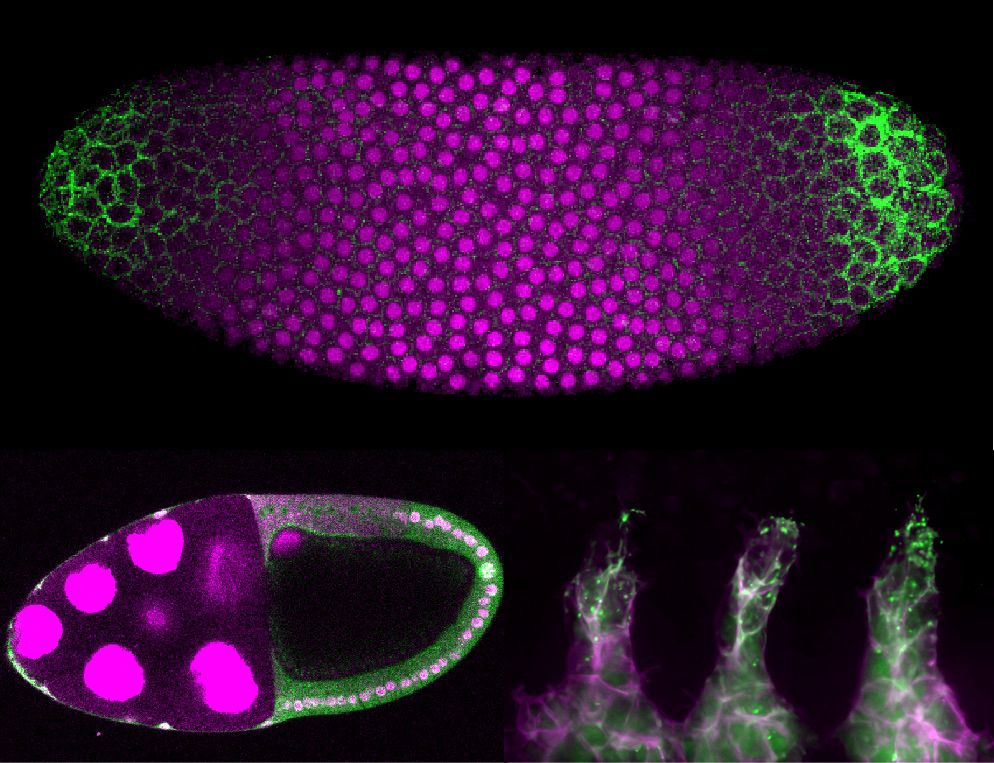
Have you ever wanted to *see* receptor activity in embryos? If so, our new preprint is for you! In it, we showcase a new live-cell biosensor for visualizing receptor tyrosine kinase activity in living embryos – pYtags!
www.biorxiv.org/content/10.1...
07.01.2025 17:45 — 👍 152 🔁 51 💬 3 📌 3

An intriguing mechanism of germline cyst formation through innovative live imaging in fetal mice gonads, and computational modeling.
Check out my dispatch in @currentbiology.bsky.social on this beautiful work.
kwnsfk27.r.eu-west-1.awstrack.me/L0/https:%2F%2…
16.12.2024 17:38 — 👍 5 🔁 1 💬 2 📌 0
Sharing my latest preprint as a re-introduction to bsky!
biorxiv.org/content/10.1...
----
Everyday, cells like this crawl through your body to protect you. While we know that their front 🔴 and back 🔵 polarity programs work together, how exactly do these two processes communicate?
11.11.2024 18:34 — 👍 36 🔁 10 💬 1 📌 0
Thank you, John! 😀
21.11.2024 03:02 — 👍 1 🔁 0 💬 0 📌 0
Thank you, Andi!
20.11.2024 22:16 — 👍 0 🔁 0 💬 0 📌 0
Can I pls be added? Thanks!
20.11.2024 19:54 — 👍 1 🔁 0 💬 0 📌 0
Please add me as well, thx! :)
20.11.2024 19:49 — 👍 1 🔁 0 💬 0 📌 0
Check out our new paper in @currentbiology.bsky.social by amazing grad students @ezlevy.bsky.social and @isabellaleite.bsky.social “A tug-of-war between germ cell motility and intercellular bridges controls germline cyst formation in mice”
authors.elsevier.com/a/1k7fg3QW8S...
20.11.2024 19:44 — 👍 89 🔁 25 💬 7 📌 5
Epigenetic (Re)Programming - Nuclear metabolism - Chromatin organisation| PhD @IMBA/@MPIB
Transcription regulation, how genes are turned on or off, transcriptional cofactors, general transcription factors and core promoters.
Evolutionary cell biology / evolution of morphogenesis / animal origins / choanoflagellates @institutpasteur.bsky.social
https://research.pasteur.fr/en/team/evolutionary-cell-biology-and-evolution-of-morphogenesis/
Stem cell biologist intrigued by the developmental functions of epigenome regulators and how they can go awry. Father of two beautiful kids. Former Tour de France contender (in his dreams). Opinions expressed here are my own.
https://www.stadtfeldlab.com
Incoming Assistant Professor, Columbia University | @DamonRunyon.org Postdoc, UC Berkeley | PhD Princeton University | Mobile elements, Structural biology and Genome engineering | http://thawanilab.org
We study receptor tyrosine kinase signaling in mammalian craniofacial development at the University of Colorado Anschutz Medical Campus. fantauzzolab.org
Postdoc at @crick.ac.uk | HFSP fellow | DevBio, epigenetics, marsupial, evo-devo | Previous Manzanares lab | #embryo2017 | (he/him)
Stem Cell Scientist @Sozen lab@Yale Genetics
Center for Regenerative Science and Medicine. UTSW. WISMAC. DevBio enthusiast. Editor of Developmental Biology. My thoughts. Hope.
https://labs.utsouthwestern.edu/cleaver-lab
Orcid: https://orcid.org/0000-0003-2454-6641
Group leader @MRC_LMB. Cell fate decisions in the early mammalian embryo
Tenure-Tracking @ NICHD_NIH
#gene_regulation #nuclear_organization #development | handkerchief user | views are mine alone, probably for the best
rochalab.nichd.nih.gov
Synthetic developmental biologist at PoL TU Dresden. Cross-species comparison and manipulation of the ORGANOID ZOO.
https://physics-of-life.tu-dresden.de/team/pol-groups/ebisuya
Quantitative Developmental Biology @Duke
Developmental biologist. Group Leader @cabd-upo-csic.bsky.social
Postdoc at Schuh lab @schuhlab.bsky.social | MPI-NAT | Microscopy • Fertility • Cell Biology
Canada Research Chair of Functional Genomics in Reproduction & Development. Associate Prof. @ Karolinksa Institutet & Université of Montreal. Everything embryos, single cell genomics, DOHaD, ART👩🏼🔬🔬🧬🧫
Geneticist. Mom of 4- & 2-legged creatures. Art enthusiast. Food lover.
Senior Editor at Nature Communications handling Developmental Biology.
Lecturer in Engineering Biology @edinburgh-uni.bsky.social
Interested in understanding and engineering cellular neighbourhoods, and coming up with funky acronyms for new synthetic biology tools.