This story started during my post-doc with @tlaszlo.bsky.social while looking at TAFs isoforms for another project. Striking that a fundamental component of the basal transcription machinery can undergo such species-specific (rather drastic) evolutionary innovations. Thanks to everyone involved!
20.01.2026 11:59 —
👍 1
🔁 1
💬 1
📌 0
Check out our preprint where we describe, characterize and trace the origin of a novel human-specific isoform of TAF8, a scaffold subunit in TFIID. We were surprised to discover that TAF8 recently underwent genetic changes generating a truncated isoform with altered properties in humans!
20.01.2026 11:59 —
👍 6
🔁 3
💬 1
📌 0
Thanks Andrea and everybody who participated!
20.01.2026 16:28 —
👍 1
🔁 0
💬 0
📌 0
A recently evolved TAF8 isoform arising from an Alu insertion increases TFIID assembly complexity in the human lineage
Despite its centrality in regulating RNA polymerase II transcription in all eukaryotes, the TFIID general transcription factor exhibits several layers of variability across different tissues and developmental stages, representing an underexplored hub of evolutionary innovation. Here, we describe a novel short isoform of TAF8 (TAF8s) — a TFIID scaffold subunit — which evolved from the use of an intronic polyadenylation site (iPAS) in the human lineage. Comparative genomics analyses show that the emergence of TAF8s expression in the human lineage coincides with minute DNA changes in the iPAS at the edge of a FLAM-C Alu element that inserted into intron 5 in the common ancestor of anthropoid primates (Simiiformes). The human-specific TAF8s isoform lacks nearly half of the canonical CDS, is widely expressed across human tissues, and constitutes a substantial fraction of the TAF8 transcript pool. TAF8s is translated into a truncated protein isoform that lacks the nuclear-localization signal and localises in the cytoplasm. TAF8s interacts with its histone-fold partner TAF10 and other core TFIID subunits, while entirely losing its interactions with TAF2, thus giving rise to alternative TFIID sub-complexes in human cells. Our study suggests that the TFIID complex underwent a recent diversification through a stepwise evolutionary acquisition of complexity in the TAF8 locus in the human lineage, leading to the expression of a novel truncated pan-isoform that might work as a modulator of TFIID assembly. We discuss the ramifications of our findings in TFIID complex diversification, its evolvability, and the genetics of TAF-related congenital disorders. ### Competing Interest Statement The authors have declared no competing interest. Fondation ARC pour la Recherche sur le Cancer, ARCPOST-DOC2021080004113 Agence Nationale de la Recherche, https://ror.org/00rbzpz17, ANR-PRCI-19-CE12-0029-01, ANR-20-CE12-0017-03, ANR-22-CE11-0013-01_ACT Fondation pour la Recherche Médicale, EQU-2021-03012631 NIH Common Fund, https://ror.org/001d55x84, R35GM139564 NSF, 1933344
Read our great story at doi.org/10.64898/2026.01.19.699933
20.01.2026 16:26 —
👍 0
🔁 0
💬 1
📌 0
High-resolution binding data of TFIID and cofactors show promoter-specific differences in vivo https://www.biorxiv.org/content/10.64898/2025.12.22.691305v1
23.12.2025 14:32 —
👍 5
🔁 3
💬 0
📌 0
Intrigued by a long-standing conundrum in small RNA biology—how nuclear Argonaute proteins silence transposons when they *need* target transcription for their own recruitment—we studied the piRNA pathway.
And found a hidden RNA-decay axis from Piwi to the RNA exosome.
22.12.2025 18:14 —
👍 98
🔁 42
💬 3
📌 5
All of this was spearheaded by the exceptional experimentalist & scientist @changweiyu.bsky.social who came with a strong background in transcription from @tlaszlo.bsky.social and developed into a remarkable RNA biologist.
22.12.2025 18:14 —
👍 6
🔁 2
💬 1
📌 0

Please RT 📧
Registration open for ➡️"Summer School on Chromatin Biology" Our 2 weeks hands on expedition 🧪👩💻August 2026. 3. edition. Learn CUT@Tag, CUT@Run, ChIPseq, ATACseq AND to analyse your own data at @helmholtzmunich.bsky.social Daily letures by experts in the field !
▶️ shorturl.at/jrA8i
09.12.2025 11:59 —
👍 11
🔁 11
💬 1
📌 0
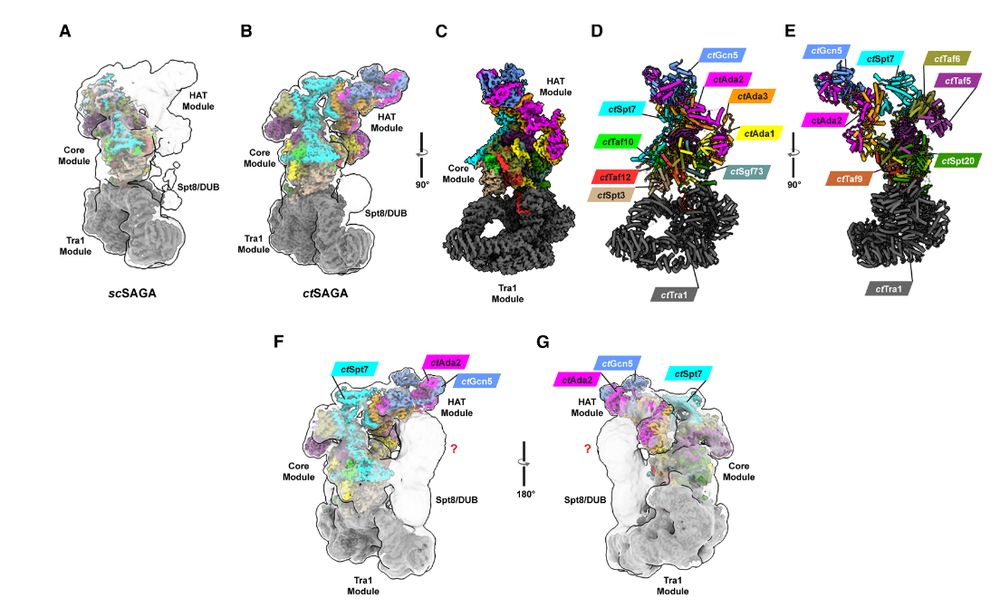
Structure of the transcriptional co-activator SAGA complex (from fungus Chaetomium thermophilum), including the histone acetyltransferase module #cryoEM
www.cell.com/molecular-ce...
21.11.2025 11:00 —
👍 12
🔁 2
💬 0
📌 0

Nobel Laureate Professor Sir John Gurdon dies aged 92
It is with great sadness that the University shares the news of the death of Professor Sir John Gurdon, founder of the Gurdon Institute.
Today it was announced that Sir John Gurdon passed away.
His ground-breaking discovery was that differentiated cells can be reprogrammed. He was a creative and distinguished developmental biologist, a kind person with a great inquisitive mind and original ideas
www.cam.ac.uk/research/new...
08.10.2025 11:56 —
👍 3
🔁 3
💬 1
📌 0

With self-deprecating humor, Sir John loved to tell the story of his Biology teacher (1949)
08.10.2025 12:10 —
👍 1
🔁 1
💬 0
📌 0
Could you add me please. Thanks. László Tora
29.08.2025 07:40 —
👍 0
🔁 0
💬 1
📌 0
One more movie!
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
29.08.2025 00:36 —
👍 46
🔁 16
💬 2
📌 0
Exquisite work. Well done to the whole team.
28.05.2025 05:33 —
👍 10
🔁 1
💬 0
📌 0
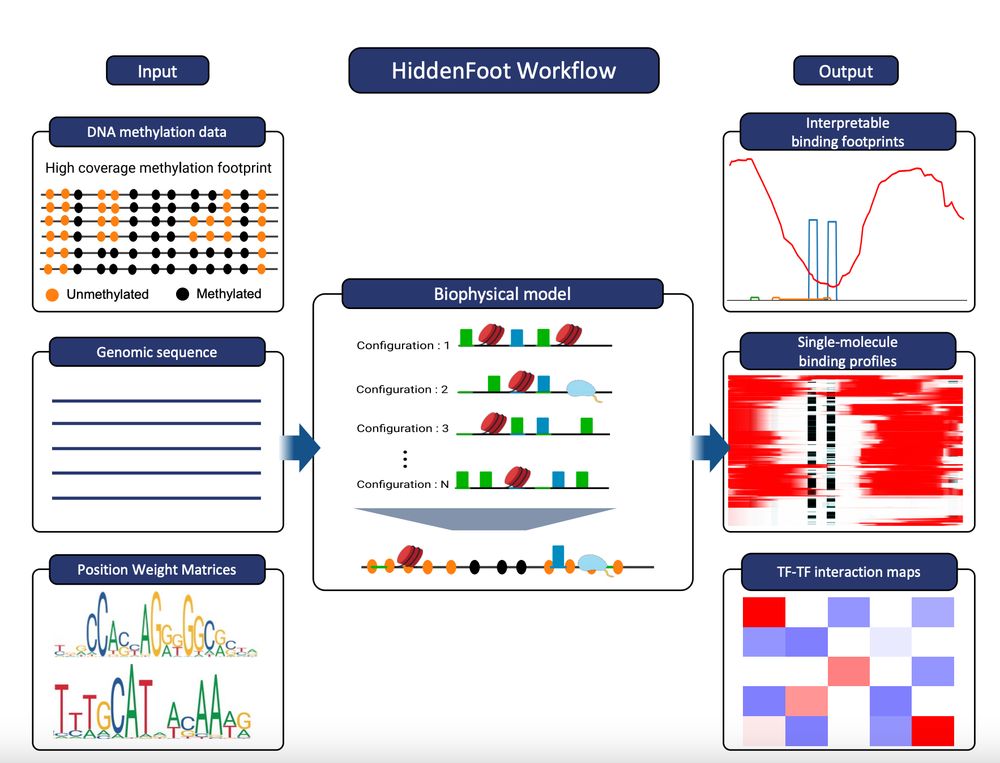
🚨 New preprint out! Do you think Single Molecule Footprinting and Fiber-seq are super cool but aren't sure how to unlock their full potential? HiddenFoot can help you: www.biorxiv.org/content/10.1...
17.05.2025 17:00 —
👍 20
🔁 5
💬 0
📌 0

Friday we celebrated my 40 years of scientific career in France, with present and past lab members and many friends. It was a fantastic day full of emotions, memories and great science. Thanks for making the day so memorable !
01.05.2025 07:08 —
👍 15
🔁 3
💬 3
📌 0
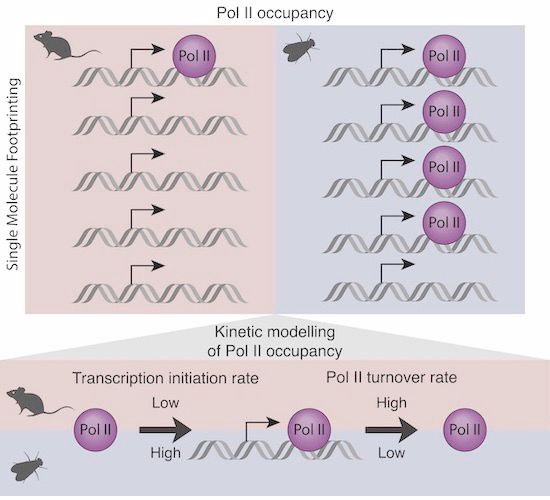
The importance of moving away from bulk! Occupancy of Pol II at promoters is dramatically different between fly and mouse cells! When looking single molecule! Proud of the team! @kasitc.bsky.social @molinalab.bsky.social doi.org/10.1038/s443...
02.04.2025 08:47 —
👍 115
🔁 37
💬 2
📌 1
Two more weeks to send us your abstract! >20 abstract selected talks. Priority to junior scientists and cool biology on gene regulation (DNA/RNA/protein). Come to meet the world leaders in single molecule genomics and single molecule imaging! Help shaping the future of the field in the workshops!
26.03.2025 12:17 —
👍 14
🔁 5
💬 0
📌 0
👍
24.03.2025 07:52 —
👍 0
🔁 0
💬 0
📌 0
PMID: 40106549, DOI: 10.1126/sciadv.adr1492
21.03.2025 17:27 —
👍 0
🔁 0
💬 0
📌 0
Read our collaborative paper
@longchrom.bsky.social and @molinalab.bsky.social
showing that disruption of NuA4 and ATAC coactivator complexes cause gene-specific transcript buffering, suggesting that cells dynamically adjust RNA export/degradation to sustain cellular homeostasis.
21.03.2025 17:27 —
👍 11
🔁 6
💬 3
📌 0













