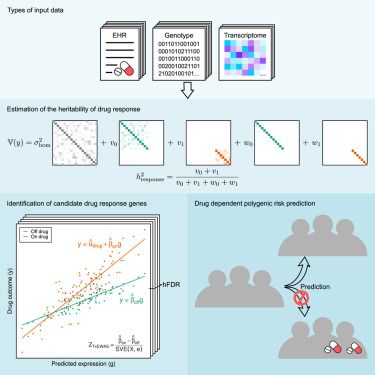
Are you using any of our factor models, such as MOFA? 🛵
You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n
07.11.2025 10:29 — 👍 34 🔁 12 💬 1 📌 3
An interesting "what have we been doing all these years?" result from this paper is how sub-optimal the widely-used uniform sampling scheme can be (cluster all @50%, sample from all clusters equally). In contrast, strategies that account for the relative differences in cluster size improve val loss
24.04.2025 07:35 — 👍 12 🔁 2 💬 1 📌 0
Single task, lightweight, short-context bp res. profile models often perform on par or outperform current large, multi task, long context models on counterfactual prediction. Much to do to improve.
Bonus: robust, efficient interpretation of syntax
Great collab with @jengreitz.bsky.social lab.
18.04.2025 14:55 — 👍 25 🔁 4 💬 0 📌 0
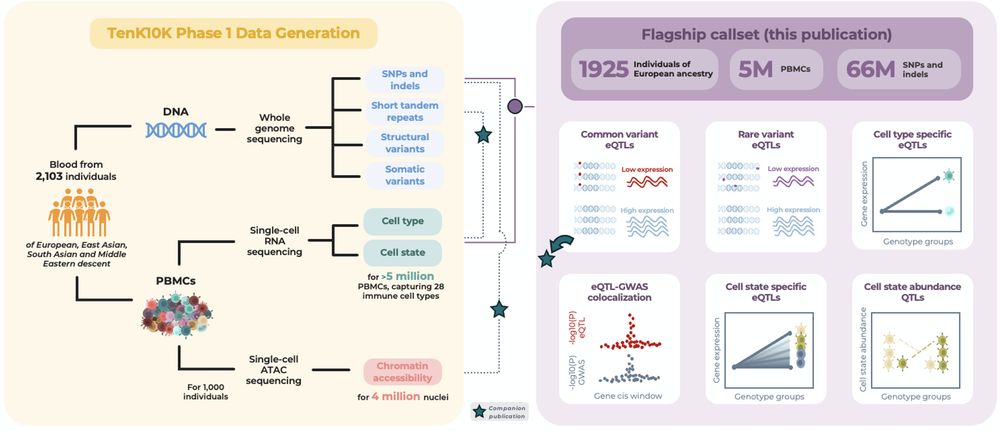
📢 new preprint alert: So so excited to share our analysis on the impact of common and rare variants on single-cell gene expression in blood, using WGS and scRNA-seq data from nearly 2,000 individuals and 5.4m cells as part of TenK10K phase 1 🧬 www.medrxiv.org/content/10.1...
🧵👇 (1/n)
24.03.2025 07:47 — 👍 99 🔁 31 💬 2 📌 1
GSK hiring Computational Biologist in Stevenage, England, United Kingdom | LinkedIn
Posted 11:13:48 PM. Site Name: UK - Hertfordshire - Stevenage, Heidelberg - OfficePosted Date: Feb 28 2025We create a…See this and similar jobs on LinkedIn.
Are you a postgraduate student interested in protein modelling and drug discovery?
We have an exciting opportunity to join our team at GSK for a 6-9 months internship, working on an ambitious cross-department research project. Apply before March 14th!
www.linkedin.com/jobs/view/41...
06.03.2025 20:23 — 👍 6 🔁 7 💬 1 📌 0
I think the regulatory model (looping etc) first is good with examples that are less extreme and unusual than fto or limb/shh. I think hmgcr and cholesterol is a proximal regulatory effect.
27.12.2024 20:42 — 👍 5 🔁 0 💬 0 📌 0
Our ChromBPNet preprint out!
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
25.12.2024 23:48 — 👍 231 🔁 89 💬 8 📌 5
We are the Stegle Lab: A bioinformatics group advancing computational methods to study molecular variations and their impact on phenotypes. We are jointly hosted at the German Cancer Research Center (@dkfz.bsky.social) and the European Molecular Biology Laboratory (@embl.org) in Heidelberg, Germany.
09.12.2024 15:24 — 👍 18 🔁 5 💬 0 📌 1
To be clear, we do have plans for scaling, we just kinda expected more than a couple days notice before getting blasted with a million new users a day.
The team is rapidly deploying fixes and new software to adapt. More servers in the mail.
20.11.2024 21:22 — 👍 38087 🔁 2942 💬 1559 📌 313
My group's work dissecting the contribution of common variants to rare neurodevelopmental conditions is now out at nature.com/articles/s41..., led by co-first authors Qinqin Huang (not yet on blue sky) and @emiliewigdor.bsky.social . See below for Emilie's tweetorial.
20.11.2024 16:15 — 👍 122 🔁 42 💬 7 📌 6

Our impact
We provide open data that helps scientists understand life and that informs solutions to real-world problems, such as infectious diseases, climate change and food security.
Not enough bioinformatics in your Bluesky feed? We’ve got you covered. Follow us for our latest news, exciting life science research, updates from our data resources, new tools and training resources.
Haven't heard of EMBL-EBI? Take a look at what we’re working on. www.ebi.ac.uk/about/our-im...
19.11.2024 14:18 — 👍 79 🔁 31 💬 0 📌 8
Hey, a question for the genetics community. Does genetic fine-mapping work well? How often does it miss?
We usually find that most fine-mapped variants do not fall within coding or regulatory regions. Is it a limitation of epigenomics or a limitation of fine-mapping? Please share your thoughts!
19.11.2024 19:56 — 👍 68 🔁 21 💬 9 📌 2
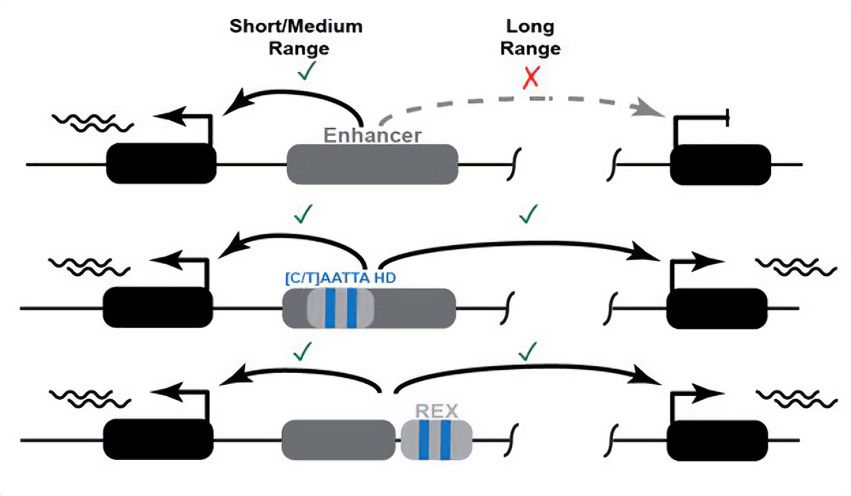
Schematic overview of the proposed mode of action of the newly discovered REX element. Top: An enhancer can activate a gene at a short distance, but not at at long range. Middle: Presence of (C/T)AATTA motifs within an enhancer enable it to act over long distances. Bottom: Coupling a short-range enhancer to the REX element containing the same motifs turns it into a long-range enhancer.
REX - a mammalian "range extender" element that can turn short-distance enhancers into long-distance enhancers.
New preprint from a collaboration led by Grace Bower and Evgeny Kvon.
doi.org/10.1101/2024...
27.05.2024 16:26 — 👍 55 🔁 21 💬 2 📌 4
Yes, and also how it informs selection of training datasets. Selecting English language text for English language comprehension, augmenting image datasets by including rotated versions of the same images for image recognition.
26.12.2023 19:40 — 👍 2 🔁 0 💬 1 📌 0
I think it already has some physics (eg covalent bond lengths), just less than you might expect (no force field concept). Similar, llms have some knowledge of language (eg the concept of words), just less than you might expect (no verb/noun concept).
26.12.2023 18:04 — 👍 1 🔁 0 💬 1 📌 0

Sad to be missing #ASHG23, but check out the talk by the brilliant Wei Zhou talk on Saturday on our new scalable & efficient method for single-cell eQTL mapping!
31.10.2023 22:46 — 👍 14 🔁 4 💬 1 📌 0
This is tremendous news, and the quote that "UK Biobank is the world’s most significant resource for health research" is not an exaggeration. Very happy to see it continue to be sustained, especially as we're doing a workshop tomorrow at #ASHG23 on how to use these data!
www.gov.uk/government/n...
31.10.2023 11:39 — 👍 10 🔁 3 💬 0 📌 0

Unfortunately I have to miss #ASHG23 this year, but if you are interested in our group's work, do check out these two posters from Ralf Tambets and Krista Freimann:
29.10.2023 13:23 — 👍 1 🔁 1 💬 1 📌 0

Presentations from our group at ASHG next week:
27.10.2023 00:25 — 👍 7 🔁 2 💬 1 📌 0

tidyomics
Open organizaion of developers creating tidy-style analysis packages in R/Bioconductor and beyond. Reach out to join. - tidyomics
Abdullah @abdnahid.bsky.social first #Bioconductor package {easylift} is released in Bioc 3.18
Facilitates genomic liftover using existing Bioc tools:
ranges |> easylift("hg38")
Abdullah is one of the group that signed up for #tidyomics open challenges: github.com/tidyomics
26.10.2023 12:54 — 👍 15 🔁 8 💬 2 📌 0

Very happy to see that our review on gene regulatory networks is being featured by Nature Reviews Genetics and that it inspired its cover! 🥳
doi.org/10.1038/s415...
16.10.2023 09:36 — 👍 21 🔁 7 💬 0 📌 1
We are doing application and collaboration driven development of interpretable and statistically sound machine learning methods for understanding disease heterogeneity
Professor of Computational Neuroscience and Psychiatry, Aarhus University. PI @ the Embodied Computation Group. We study perception, interoception, & metacogniton.
https://www.the-ecg.org
Computational Biophysicist; Physics Professor & Physics Graduate Director at WFU; Editor-in-Chief, Journal of Biomolecular Structure & Dynamics
Genetics and Epigenetics of Cardio Metabolic Traits. Oxford.
Research collaborator @ Imperial College London, visiting scientist @ Broad Institute.
Writer | Editor | Medical Journalist | Biopharma Analyst | Singer
PhD, Statistical Geneticist @ Genomics
Endocrinologist. Physican scientist. Director of MRC Metabolic Diseases Unit, University of Cambridge. Expressing my own opinions here, not my employers
We enable open science by developing and implementing
open source solutions, FAIRifying data, and offering expert consulting in life sciences.
Learn more: www.thehyve.nl
Associate Dean for data science, and Professor of Medicine and Human Genetics at the University of Chicago.
Academy Professor in computational Bayesian modeling at Aalto University, Finland. Bayesian Data Analysis 3rd ed, Regression and Other Stories, and Active Statistics co-author. #mcmc_stan and #arviz developer.
Web page https://users.aalto.fi/~ave/
Switchy leatherman into bad puns, thick boys, and distributed systems. Purveyor of fine jockstrap selfies. Woof.group admin.
Jim Starkey: "Of interest only to third-rate […]
🌉 bridged from ⁂ https://woof.group/@aphyr, follow @ap.brid.gy to interact
Associate professor at the Hebrew University of Jerusalem.
Statistical, population, and medical genetics; preimplantation genetic testing. Views my own.
http://scarmilab.org
Dataframes powered by a multithreaded, vectorized query engine, written in Rust.
Research & code: Research director @inria
►Data, Health, & Computer science
►Python coder, (co)founder of scikit-learn, joblib, & @probabl.bsky.social
►Sometimes does art photography
►Physics PhD
🧪 Data science, survey science, social science
💻 Director of Data Science @ Microsoft Garage
[Posts do not represent my employer]
🧮 Stats, R, python
📝 Science, Research: measurement, social biases, emotion. Ex-academic but scientist at heart
Head, Dept of Biochemistry & Molecular Genetics, Univ of Illinois, College of Medicine: cell biology, vascular biology, genomics, computational biology, immunology, regeneration, neuroscience; https://chicago.medicine.uic.edu/bmg/profiles/rehman-jalees/
Our group develops and applies computational approaches to study molecular variations and their phenotypic consequence. We are part of DKFZ and EMBL.
Website: https://steglelab.org/
Mentor, scientist & engineer. Having fun in @slavovlab.bsky.social and Parallel Squared Technology Institute @parallelsq.bsky.social with biology & single-cell proteomics.
https://nikolai.slavovlab.net