Meet the keynote speakers for the 2025 scverse conference!
Erika Alden DeBenedictis, Co-founder of The Align Foundation 🧵
@erika-alden.bsky.social
@alignbio.bsky.social
@pioneerlabs.bsky.social
#scverse #scverse2025 #StructuralBiology #Biotech #Stanford
04.11.2025 16:37 — 👍 3 🔁 1 💬 1 📌 0
"Usability besides performance is also key."
Cannot stress this enough. There's a ton of methods out there that might not even be the best but are used because a substantial amount of time was invested into making it fast, accessible, and documented. Such methods are benchmarked in real use cases
24.10.2025 13:12 — 👍 4 🔁 0 💬 0 📌 0


🌟 Thank You to Our Platinum Sponsor: 10x Genomics! 🌟
We're delighted to recognize 10x Genomics as a Platinum Sponsor of scverse conference 2025! 🏆
@10xgenomics.bsky.social
#scverse2025 #ComputationalBiology #SingleCell #SpatialOmics #10xGenomics #Bioinformatics #Biotechnology
16.10.2025 15:11 — 👍 1 🔁 1 💬 1 📌 0
Hi bioinformatics, genomics and CS friends! Please help me spread the word. I'm hiring a postdoc! Come work on cutting edge method development in algorithmic genomics with me and my group at @umdscience.bsky.social! 🖥️🧬
10.10.2025 13:02 — 👍 29 🔁 37 💬 0 📌 3

My favorite part of last-year #scverse conference was to finally meet in-person many developers that I only knew from their GitHub username!
Submissions and travel grant applications close in 3 days. Find out more and apply at scverse.org/conference2025
@scverse.bsky.social
12.09.2025 20:11 — 👍 15 🔁 5 💬 0 📌 0
scVerse conference is at Stanford this year! Encourage folks (especially Bay Area/West coast folks) to sign up and attend. Cool workshops/talks and other events + great community of developers for one of the most widely used open source software for single cell data analysis!
26.08.2025 18:50 — 👍 20 🔁 9 💬 0 📌 1
It is pretty heart warming to see people start their really good pull request with "this is my first open source contribution. I hope that this is good.". Everyone starts somewhere and we're always happy to guide at @scverse.bsky.social .
18.08.2025 20:31 — 👍 1 🔁 0 💬 0 📌 0

Our benchmark + guidelines for atlas-level differential gene expression of single cells is online:
academic.oup.com/bib/article/...
Bottom line: Use pseudobulk + DESeq2 in simple and pseudobulk + DREAM in more complex settings.
Collab w/ @leonhafner.bsky.social @itisalist.bsky.social
13.08.2025 05:51 — 👍 15 🔁 6 💬 1 📌 0
I hate reference limits of journals so much. I'm sitting here trying to find a few more references to remove despite me knowing that I'll either remove context & evidence for scientific conclusions or not mention tools that very much deserve to be mentioned and cited.
08.08.2025 17:56 — 👍 2 🔁 0 💬 0 📌 0
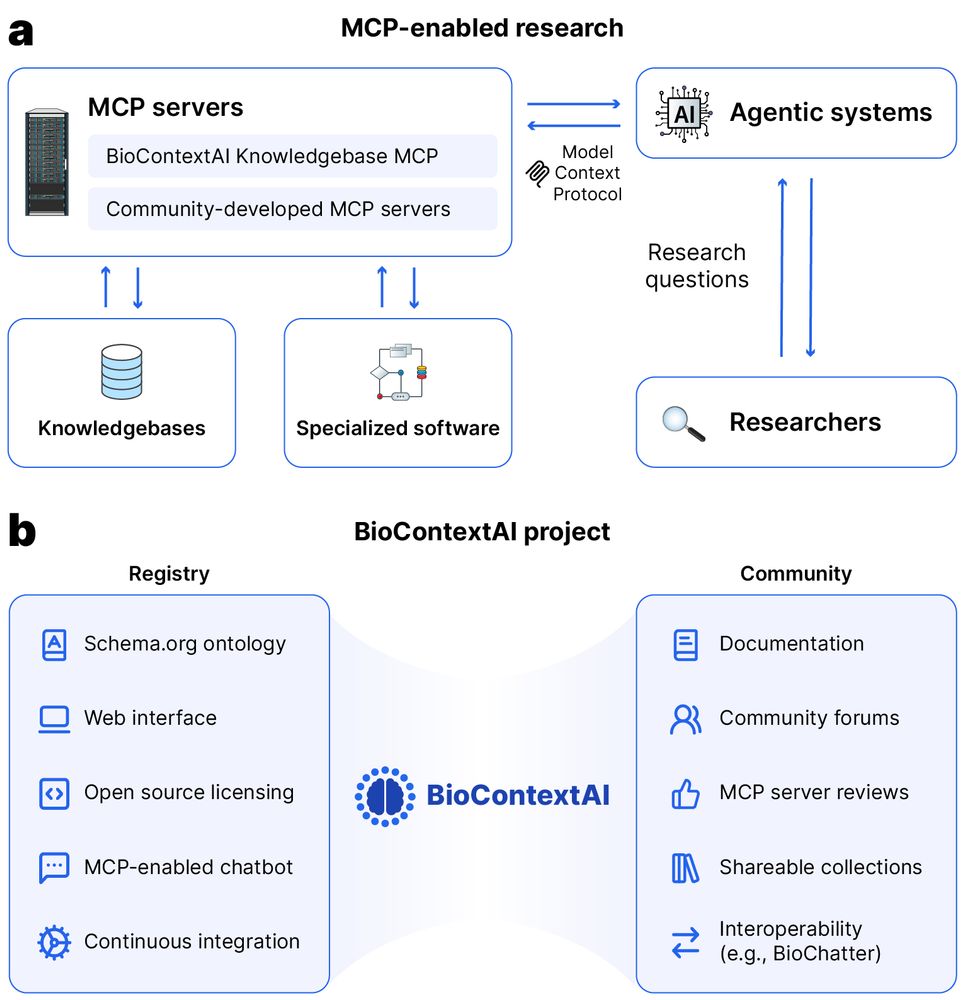
Diagram showing MCP-enabled research architecture. Part A illustrates the flow between MCP servers (including BioContextAI Knowledgebase MCP and Community-developed MCP servers), Agentic systems connected via Model Context Protocol, and researchers asking research questions. MCP servers connect to knowledgebases and specialized software. Part B details the BioContextAI project components, divided into Registry (Schema.org ontology, Web interface, Open source licensing, MCP-enabled chatbot, Continuous integration) and Community (Documentation, Community forums, MCP server reviews, Shareable collections, Interoperability with tools like BioChatter).
Preprint alert 🚨 Do you use chatbots in your work or even build MCP servers and agentic systems yourself?
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
29.07.2025 12:41 — 👍 14 🔁 10 💬 1 📌 2
Happy to have this finally out! Check out the updated vignettes and if something is broken please open a GitHub issue and I'll have a look 😉👇
12.06.2025 11:32 — 👍 9 🔁 1 💬 0 📌 0
GitHub - quadbio/cell-annotator: Automatically annotate cell types, consistently across samples.
Automatically annotate cell types, consistently across samples. - quadbio/cell-annotator
Happy to share CellAnnotator, a lightweight Python package to query OpenAI models for initial cell type annotations: github.com/quadbio/cell...
Inspired by ideas in www.nature.com/articles/s41... and github.com/VPetukhov/GP..., implemented as an scVerse ecosystem package with docs and tutorials.
28.03.2025 16:53 — 👍 8 🔁 4 💬 1 📌 0
GitHub - theislab/multimil: Multimodal weakly supervised learning to identify disease-specific changes in single-cell atlases
Multimodal weakly supervised learning to identify disease-specific changes in single-cell atlases - theislab/multimil
Tomorrow at 2025-04-15 18:00 CEST, we’ll have our next community meeting!
Anastasia Litinetskaya will present “Multimodal integration and identification of disease-specific changes in single-cell atlases”. Check out the GitHub repo here: github.com/theislab/mul...
14.04.2025 22:41 — 👍 6 🔁 1 💬 1 📌 1
We are back 🔥
14.04.2025 22:45 — 👍 2 🔁 1 💬 0 📌 0

Release notes
Version 1.11: 1.11.0 2025-02-14: Release candidates: rc2 2025-01-24, rc1 2024-12-20. Features: rc1 sample() supports both upsampling and downsampling of observations and variables. subsample() is n...
🎉 Scanpy 1.11.0 is out! 🎉 just after reaching 2000 stars on GitHub!
- sc.pp.sample replaces subsample with many new features
- Sparse Dask support pca
- session-info2 package for more reproducible notebooks
See the release notes:
14.02.2025 12:08 — 👍 49 🔁 19 💬 1 📌 1
We have additional available spots. Please consider joining! It will be a blast.
30.01.2025 16:24 — 👍 2 🔁 2 💬 0 📌 0
I keep being astonished at how terrible @microsoft.com Teams works on Firefox and I certainly don't blame Mozilla for it.
22.01.2025 09:46 — 👍 1 🔁 0 💬 0 📌 0
Why is there still no proper "thank you" emoji on @github.com ? I am grateful for other peoples time and feel like a thumbs up is kind of like a drive by reaction and a heart isn't always perfectly appropriate
14.01.2025 12:51 — 👍 3 🔁 0 💬 1 📌 0
I actually think that it' awesome that even fundamental packages like numpy, pandas, scipy, and others still develop rapidly and aren't afraid of making big API changes but it's quite time consuming to keep up when one maintains many packages. I can't imagine what e.g. the numba devs go through
10.01.2025 16:57 — 👍 3 🔁 0 💬 0 📌 0
Awesome, congratulations! It was a pleasure to attend Yimin's talk at the @scverse.bsky.social 2024 conference and it's really nice to see that it was encouraging for Yimin.
07.01.2025 15:26 — 👍 1 🔁 0 💬 1 📌 0
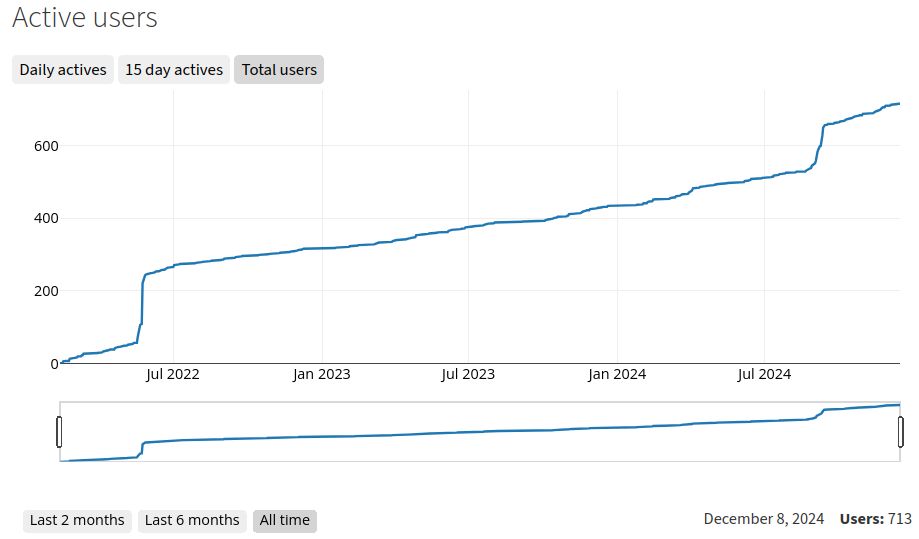
I was just looking at our @scverse.bsky.social zulip chat statistics and am super happy about our steady and healthy growth. The two bursts are our official launch in May 2022 and the inaugural scverse conference in September of 2024. Please join us! scverse.zulipchat.com
15.12.2024 15:52 — 👍 12 🔁 6 💬 0 📌 0
Reminder!Today at 18:00 CET will be the last community meeting of the year! @lukasheumos.bsky.social will talk about about pertpy, a Python-based modular framework for the analysis of large-scale perturbation single-cell experiments. Zoom link can be found here: scverse.zulipchat.com#narrow/chann...
10.12.2024 12:24 — 👍 2 🔁 2 💬 0 📌 0
GitHub - scverse/pertpy: Perturbation Analysis in the scverse ecosystem.
Perturbation Analysis in the scverse ecosystem. Contribute to scverse/pertpy development by creating an account on GitHub.
Next Tuesday at 2024-12-10 18:00 CET, we will hold the last community meeting of the year!
@lukasheumos.bsky.social will talk about pertpy, a Python-based modular framework for the analysis of large-scale perturbation single-cell experiments. The GitHub repo: github.com/scverse/pertpy
03.12.2024 09:56 — 👍 25 🔁 5 💬 1 📌 0

If this message does not go away, try reloading the page.
If this message does not go away, try reloading the page.
Reminder! Today at 18:00 CET will be another community meeting! Malte Lücken will talk about about Open Problems in Single-Cell Analysis. Zoom link can be found here:
26.11.2024 08:31 — 👍 9 🔁 4 💬 2 📌 0
We were thinking of keeping that starter pack only for scverse core team and board members. There's already other pure single-cell lists and we don't want to compete with them. We don't need several lists that have the same purpose. Thank you very much for your interest though!
25.11.2024 18:30 — 👍 1 🔁 0 💬 0 📌 0
Computational Biologist @Merck KGaA Darmstadt, DKFZ alumni. data-driven insights
PhD student at the Yosef Lab at the Weizmann Institute of Science | Computational Biologist
PostDoc @mpibiochem.bsky.social in @mannlab.bsky.social | PhD in Biochemistry | Developer of scPortrait & SPARCS | DL-driven representation learning for single-cell image analysis
https://sophia-maedler.com
Community initiative for biomedical MCP servers 🤖🧬
Automatically shares new MCP servers from the Registry
Find out more at: https://biocontext.ai
PhD student at Stanford CS; AI+Biomedicine
kexinhuang.com
Assistant Professor in the Section of Computational Biology, Division of Oncology, Dept. of Medicine at WashU
scverse Core Member & Community Manager
Evans MDS Young Investigator
jfoltzlab.org
PhD student @ Helmholtz Munich | Interested in Genetics, Omics, Digital Health, Deep Learning.
Assistant Professor for Data Science in Systems Biology at the Technical University of Munich (http://daisybio.de). Mostly posting about bioinformatics and systems / network biology research. Views are my own. he / him.
I am a Bioinformatician whose research focusses on post-transcriptional regulation in the context of Alzheimer's Disease.
---
https://mschilli.com
trying to help patients by understanding their individual immune status
http://scholar.google.com/citations?user…
Doing a Ph.D. AI in Bio. | Ex @WhiteLabGx @BroadInstitute @MIT | Built @PiPleteam | ML, Cancer, Genomics, Data Sci, Entrepreneur, FullStack Dev | All views are mine
Computer Science PhD Student at @humboldtuni.bsky.social and @mdc-berlin.bsky.social | Data Science | Machine learning | AI | Bioinformatics | Genomics | Single-Cell Biology
Research Scientist AI/ML @cziscience.bsky.social , PhD @www.helmholtz-munich.de , OSS @scverse.bsky.social
giovannipalla.com
Married to @outlawren.bsky.social ❤️
from Milwaukee, Wisconsin, USA
Bad River Ojibwe
Principal Investigator at CeMM and LBI-NetMed: https://rendeiro.group
Computational Biology of Aging and Pathology
PhD student in machine learning and comp bio at the Fabian Theis lab, Helmholtz Munich. Interests: single cell, ML, cancer, atlases, ML in the clinic, philosophy, and 💃
Our group develops and applies computational approaches to study molecular variations and their phenotypic consequence. We are part of DKFZ and EMBL.
Website: https://steglelab.org/
Doctoral Researcher specializing in "Machine Learning and Computational Biology" at TUM & Helmholtz, part of Fabian Theis research group.
kemalinecik.com