GitHub - LArnoldt/networkvi
Contribute to LArnoldt/networkvi development by creating an account on GitHub.
NetworkVI is a group effort by @juliusuzb.bsky.social, Luis Herrmann, Khue Nguyen, supervised by @fabiantheis.bsky.social & Benjamin Wild & Roland Eils at @bihatcharite.bsky.social and @www.helmholtz-munich.de.
Code: github.com/LArnoldt/networkVI
🧠 Open-source & ready to use!
16.06.2025 07:09 — 👍 3 🔁 2 💬 0 📌 0
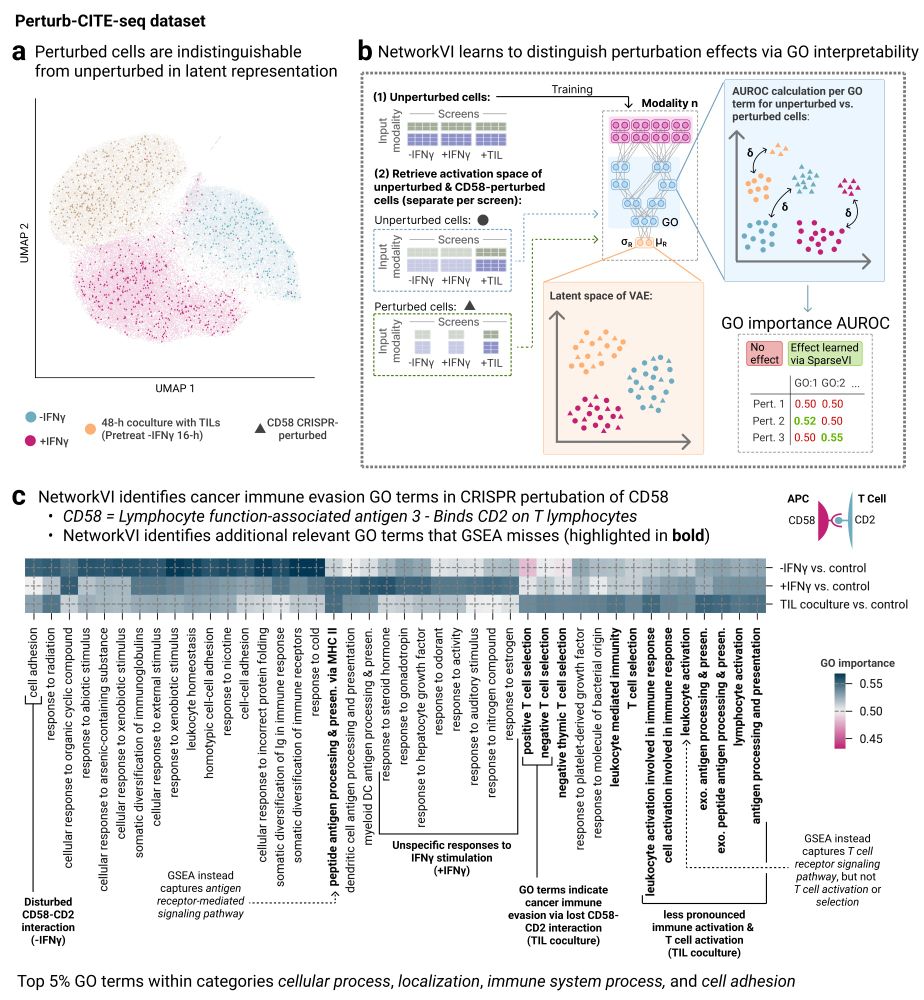
We demonstrate how NetworkVI identifies immune evasion mechanisms via GO programs in a CRISPR-perturbed melanoma dataset not detectable with standard methods. This highlights the medical utility of interpretable multimodal models.
16.06.2025 07:05 — 👍 0 🔁 0 💬 1 📌 0
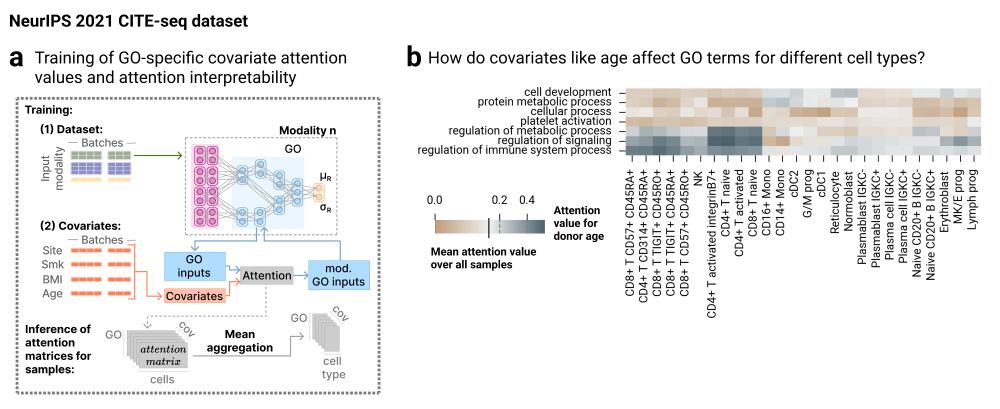
NetworkVI also incorporates a GO-specific covariate attention mechanism, allowing the model to adjust for sample-level metadata (e.g., donor or condition), improving both performance and interpretability in heterogeneous datasets.
16.06.2025 07:05 — 👍 0 🔁 0 💬 1 📌 0
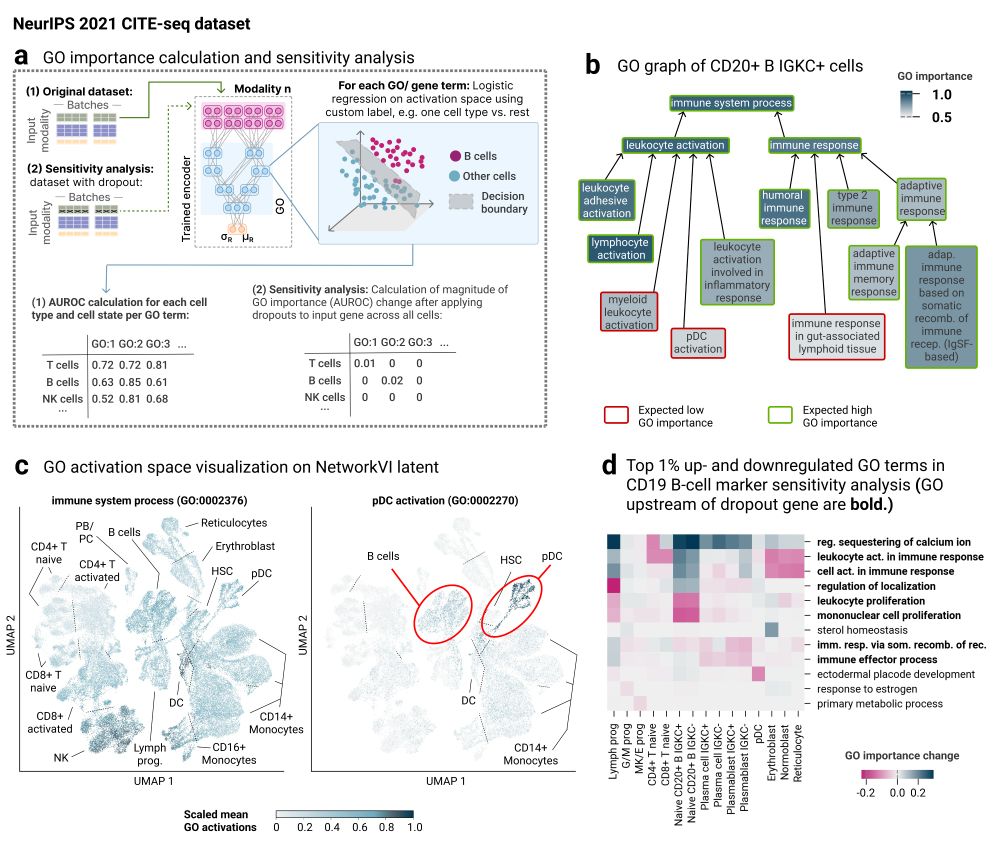
These scores reveal associations between genes and GO terms, helping to identify active biological processes and their regulatory drivers in a given condition or perturbation.
This opens a path toward mechanistic interpretation beyond enrichment tests.
16.06.2025 07:04 — 👍 0 🔁 0 💬 1 📌 0
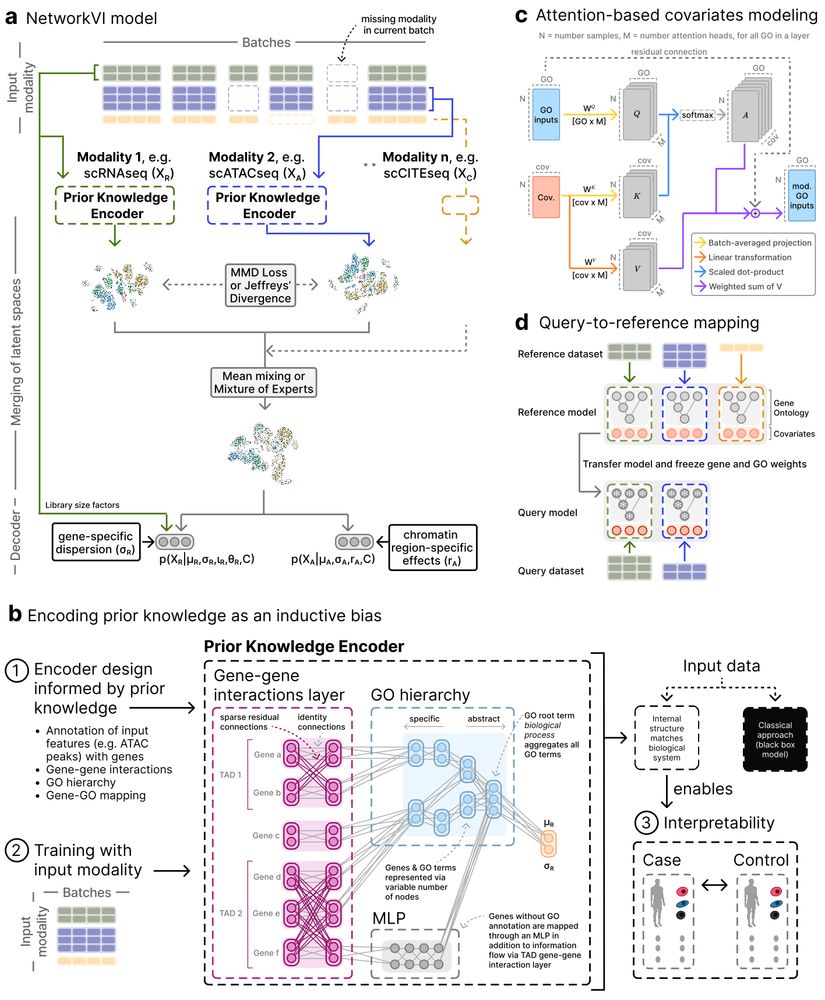
NetworkVI’s architecture incorporates prior biological knowledge:
1. Gene-gene interactions from TADs
2. The structure of the Gene Ontology
This enables inference of gene and GO term importance scores at modality- and cell-specific resolution.
16.06.2025 07:03 — 👍 0 🔁 0 💬 1 📌 0
📊 NetworkVI achieves state-of-the-art performance on bimodal and trimodal data:
✅ Accurate integration
✅ Imputation of missing modalities
✅ Query-to-reference mapping
✅ Interpretation of cell type– and perturbation-specific signatures
16.06.2025 07:02 — 👍 0 🔁 0 💬 1 📌 0
Current multimodal single-cell integration methods act as black boxes, lacking meaningful interpretability. We introduce NetworkVI, a VAE that performs integration via gene-gene interactions and the Gene Ontology for biologically grounded analysis. www.biorxiv.org/content/10.1...
16.06.2025 07:02 — 👍 1 🔁 0 💬 1 📌 0
PhD student @yalecbb.bsky.social in the lab of @hattaca.bsky.social | interested in single-cell genomics, drug discovery, and AI | former data analyst at Northwestern PCCM | alum of WashU BME
https://sites.google.com/yale.edu/samfenske
Pediatric Brain Cancer Researcher focused on DIPG | Postdoc Vitanza Lab | PhD on DIPG Australia | MSc on GBM UK | BSc on GBM Germany | Passionate runner, cyclist and swimmer
Clinician Scientist I Neuropathology @ University Clinic of Freiburg | Hem/Onc - Neuroimmunology - Precision medicine | EANO Youngster Committee
PhD Student in Computational Biology at TU Munich (Gagneur lab) and Helmholtz Munich (Theis lab)
Interested in rare variants and their effect in Population-scale cohorts
Associate Professor of Bioinformatics at University of Tartu, Estonia. Project lead at eQTL Catalogue. https://kauralasoo.github.io/
feeds: biorxiv, medrxiv, arxiv.q-bio, arxiv.cs, arxiv.math, Research Square
regex: '(single(- )(cell|nuclei)|s[cn]RNA-seq)'
If you have suggestions, please get in contact with @willmacnair :)
Bioinformatics Scientist / Next Generation Sequencing, Single Cell and Spatial Biology, Next Generation Proteomics, Liquid Biopsy, SynBio, Compute Acceleration in biotech // http://albertvilella.substack.com
Translating the Science of Health & Longevity
USD Bio Professor, Drug Developer
📖 Longer content on LinkedIn:
linkedin.com/in/jon-brudvig-phd-9b1830134
📰Sign up for my monthly Substack newsletter, Translation:
jonbrudvig.substack.com
physician-scientist, author, editor
https://www.scripps.edu/faculty/topol/
Ground Truths https://erictopol.substack.com
SUPER AGERS https://www.simonandschuster.com/books/Super-Agers/Eric-Topol/9781668067666
Foundational tools for omics data (mostly in python).
Core Developer @scverse & Solutions Architect @NVIDIA
Assistant Professor leading the Computational Biomedicine Group at the University of Innsbruck • Data scientist at the Digital Science Center (DiSC) • Precision medicine and cancer immunotherapy • Mother 🧬🖥️
https://computationalbiomedicinegroup.github.io/
Single Cell/Spatial. Cancer Immunology. Outdoor activities.
Core developer @scverse.bsky.social.
Working in Clinical Bioinformatics at Boehringer Ingelheim.
Formerly PhD student at Medical University of Innsbruck.
My private account.
github.com/grst
Computing all things single-cell | Postdoc at Genentech @MarioniLab | Steering council at scverse.org
Research software engineer at Lamin Labs | Steering council at scverse | Postdoc at Fabian Theis lab
Postdoc fellow @ Stanford & Gladstone Institutes
Core team @scverse-team.bsky.social
Bringing the single-cell genomics in human complex trait genetics
https://emdann.github.io/
PhD brain and genomic scientist. Ocean junkie. Computer wrangler. Open data believer. Geek. Punk. Not that kind of doctor. Views only my own. She/her
Associate Professor of Data Science & Dean of Research at UVA School of Data Science, #Rstats enthusiast, dad, runner, guitar noise-maker. Views my own.
Web: https://stephenturner.us/
Newsletter: https://blog.stephenturner.us



