GitHub - Benoitdw/warnme: Library / CLI to communicate with me
Library / CLI to communicate with me. Contribute to Benoitdw/warnme development by creating an account on GitHub.
It's easy to receive an email when a job finishes on a computer cluster, but setting that up on a single machine is a little less straightforward. Thanks to my colleague Benoît I discovered you can very easily send yourself messages via a Telegram bot from Python!
github.com/Benoitdw/war...
18.10.2025 12:21 — 👍 1 🔁 0 💬 0 📌 0

GitHub - pola-rs/polars: Dataframes powered by a multithreaded, vectorized query engine, written in Rust
Dataframes powered by a multithreaded, vectorized query engine, written in Rust - pola-rs/polars
This week I discovered the python library polars, it's like pandas but based on Rust so it's much faster. I was working with a big dataframe (around 100 million rows) and I was blown away by its speed. Highly recommend you try it!
github.com/pola-rs/polars
06.09.2025 16:40 — 👍 1 🔁 0 💬 0 📌 0
A simple flow of gas over water may have helped kickstart life on early Earth.
Research shows how evaporation and fluid flow in a narrow channel can drive isothermal DNA replication, offering a plausible, low-temp setting for early molecular evolution.
buff.ly/ADgyeFk
24.07.2025 22:28 — 👍 15 🔁 5 💬 0 📌 1
Matsvei Tsishyn will also be there at poster A-063 to present our work (mostly his): StructureDCA, "Inferring Protein Mutational Landscape with Structure-Informed Direct Coupling Analysis"
#ISMBECCB2025 #3DSIG
20.07.2025 21:14 — 👍 0 🔁 0 💬 0 📌 0
See you tomorrow, poster B-443! #ISMBECCB2025
20.07.2025 21:10 — 👍 0 🔁 0 💬 1 📌 0
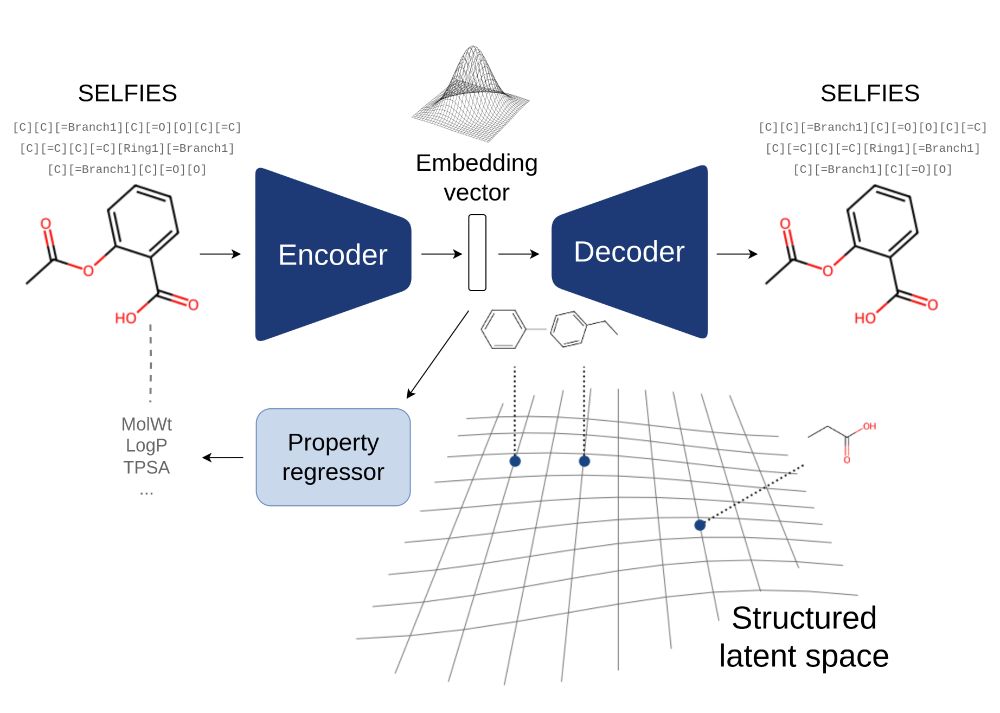
Graphical abstract of our VAE. The encoder inputs a SELFIES string representing a molecule and encodes into an embedding vector in a structured latent space. The decoder can decode this embedding vector into the original molecule.
Two weeks from now I'll be at ISMB 2025 in Liverpool with a poster presenting our Transformer-based small molecule VAE with a chemistry-aware latent space. In the meantime, if you're attending, you can already see it online: iscb.junolive.co/ISMB/live/ex...
Looking forward to seeing you there!
08.07.2025 12:00 — 👍 2 🔁 0 💬 1 📌 0
1/n
Looking through this. Will pull tidbits out and put them in this thread.
30.05.2025 23:39 — 👍 2 🔁 3 💬 1 📌 0
NIH funding supporting the HMMER and Infernal software projects has been terminated. NIH states that our work, as well as all other federally funded research at Harvard, is of no benefit to the US.
22.05.2025 12:42 — 👍 287 🔁 232 💬 37 📌 46
And that's the goal of my current research: design new molecules that target some specific olfactory receptors.
And that's a challenge... more on that later! :)
24.05.2025 13:44 — 👍 0 🔁 0 💬 0 📌 0
Interestingly, olfactory receptors aren’t limited to the nose: they’re also expressed in various tissues where they regulate different functions (blood pressure, glucose metabolism, hair growth, even play a role in prostate cancer...)...
which makes them promising drug targets!
24.05.2025 13:44 — 👍 0 🔁 0 💬 1 📌 0
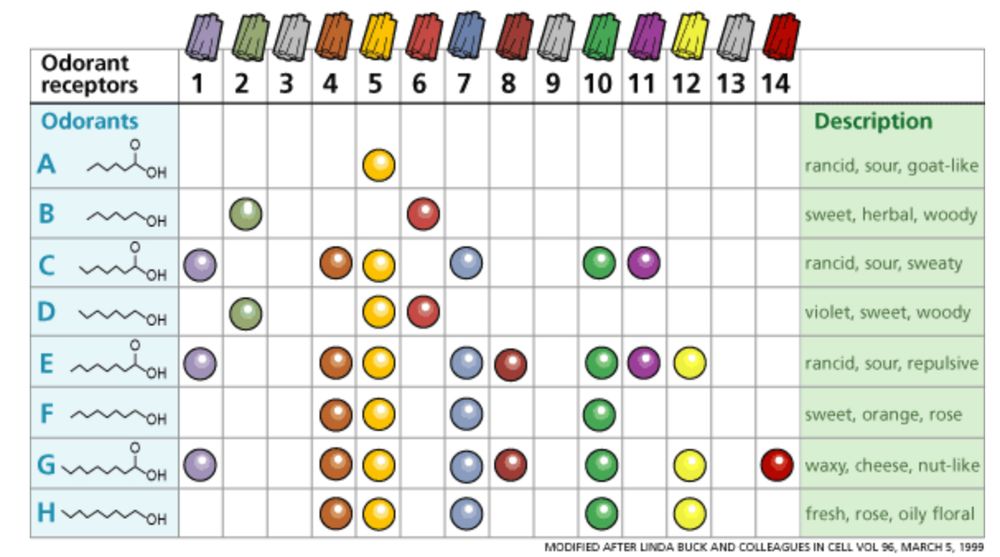
Illustration of the olfactory combinatorial code, by C. Trimmer, J.D. Mainland, in "Chapter 17 - The Olfactory System", 2017
Olfaction works like a combinatorial code: there are various types of olfactory receptors, each can be activated by multiple molecules, and some molecules can activate multiple receptors. The brain interprets these unique activation patterns as distinct smells.
24.05.2025 13:44 — 👍 0 🔁 0 💬 1 📌 0
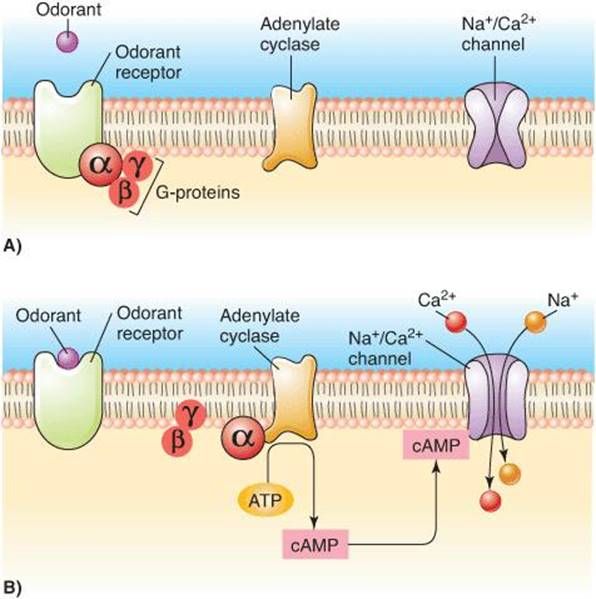
Olfactory receptor activation mechanism.
Figure from https://doctorlib.org/physiology/ganong-review-medical-physiology/13.html
Did you know smell detection relies on specialized proteins called "olfactory receptors"?
Located in olfactory neuron membranes inside the nose, they change shape when a molecule binds, triggering reactions that ultimately open ion channels, depolarize the membrane and send a signal to the brain.
24.05.2025 13:44 — 👍 0 🔁 0 💬 1 📌 0
Hi! I'm a little late to the party, but glad to be finally joining BlueSky! I'll be posting a little bit about the work I do as a postdoctoral researcher at ULB, also probably random fun facts I learn here and there, because biology never ceases to amaze me.
18.05.2025 18:11 — 👍 4 🔁 0 💬 0 📌 0
A fully open access, peer-reviewed journal published jointly by Oxford University Press and the International Society for Computational Biology.
Enzyme mechanisms, Structural Biology, Microbiome - Université Paris-Saclay, @univparissaclay.bsky.social, INRAE, @inrae-france.bsky.social, MICALIS institue, ChemSyBio Laboratory
https://www.micalis.fr/equipe/chemsybio/
Head of Department of Algorithmics and Software,
Silesian University of Technology
bioinformatician, computer scientist
Décrypter la complexité et la diversité du vivant
🔗 https://www.insb.cnrs.fr/fr
Bienvenue sur le compte d'INRAE
Institut national de recherche pour l’agriculture, l’alimentation et l’environnement
Association des Jeunes Bioinformaticiens de France - Antenne Française de l'ISCB SC
jebif.fr
Professor of Chemistry, University of Bristol. Computational chemistry,enzyme catalysis, biomolecular simulation,HPC,antibiotic resistance.Views my own.
The Ensembl project seeks to enable genomic science by providing high-quality, integrated annotation.
Vertebrates: www.ensembl.org
Non-vertebrates: www.ensemblgenomes.org
You can test the new Ensembl browser and share your feedback at beta.ensembl.org
How are biological forms encoded? Group leader at IBDM Marseille, director of Turing Center for living systems (CENTURI). Professor at Collège de France, Paris: Dynamics of living systems
The SIB Swiss Institute of Bioinformatics is an internationally recognized non-profit organization dedicated to biological and biomedical #DataScience.
We provide essential resources, expertise and services and represent Swiss bioinformatics.
Research Fellow in Bioinformatics (Proteins + ML + Function) @ CATH University College London | Mountains, Proteins, Food in that order | MSCA Alumn | Replies to emails!
Posts mine alone, in English (mostly) & French. #Evolution #Bioinformatics #openaccess #EDI. Director of @dee_unil and group leader at @SIB. He/Him. PI of @bgeedb […]
[bridged from https://ecoevo.social/@marcrr on the fediverse by https://fed.brid.gy/ ]
Prof. of Bioinformatics at Dundee, Scotland, UK and head of https://bartongroup.org the home of @jalview.bsky.social jalview.org JPred and more! See: https://bit.ly/3YXtgeq for the publications.
Also writes music: https://youtube.com/@GeoffBartonMusic
News from Geoff Barton's Computational Biology Research Group.
Home of Jalview https://www.jalview.org, JPred and much more! See: https://bit.ly/3YXtgeq for the publications.
https://bartongroup.org
https://jalview.org
Also see: @gjbarton.bsky.social
The #OpenScience journal from BGI & OUP publishing articles using/generating large datasets. And linked to GigaDB #opendata hosting/analysis repository.
Website: http://gigasciencejournal.com
Empowering Computational Biology, Transforming Global Science!
The International Society for Computational Biology is a scholarly society for advancing understanding of living systems through computation, and communicating scientific advances worldwide.
Bioinformatics Research Associate at Northwestern Medicine Chicago | Urologic Cancers @ Yang Lab | Transcript Discovery | Exploring Genomic Alterations | Neoantigens and Immunotherapy
Réseau Stand For Science Paris et région parisienne.
Les sciences sont un bien commun !
#StandUpForScience
Site national : https://standupforscience.fr/
Postdoc at NYU CDS and Flatiron CCN. Wants to understand why deep learning works.