Last call! This is a great opportunity for those interested in learning and applying SOTA ML/DL models to microscopy data with the leading experts in the field.
Personally super excited to be a lead TA along with the amazing @afoix.bsky.social and @edhirata.bsky.social!
Hope to see you there :)
15.01.2026 10:22 — 👍 8 🔁 4 💬 0 📌 0

Now speaking #CBIAS2025 @albertdm.bsky.social from @maweigert.bsky.social introducing spotiflow www.nature.com/articles/s41...
24.11.2025 11:15 — 👍 10 🔁 3 💬 1 📌 0
Had a blast at #CBIAS2025! Great science (like the incredible ShapeEmbed talk from @afoix.bsky.social), amazing people and obviously tons of beautiful images :) Already looking forward to coming back next year!
25.11.2025 23:33 — 👍 8 🔁 1 💬 0 📌 0

We’ve upgraded ShapeEmbed 🎉 ShapeEmbedLite decodes latent codes via an MLP to guarantee valid EDMs, making it lighter and ideal for small microscopy datasets or limited compute. Hear more at my BIC workshop talk or poster at #ICCV2025! Try it out at github.com/uhlmanngroup...
17.10.2025 07:35 — 👍 15 🔁 5 💬 1 📌 1

🧠 The Lipid #Brain Atlas is out now! If you think #lipids are boring and membranes are all the same, prepare to be surprised. Led by @lucafusarbassini.bsky.social with Giovanni D'Angelo's lab, we mapped membrane lipids in the mouse brain at high resolution.
www.biorxiv.org/cgi/content/...
16.10.2025 06:23 — 👍 282 🔁 110 💬 7 📌 11
You may have heard me talk about learned shape representations for a long time, but it took @afoix.bsky.social's hard work to bring it to life 🍾 Catch her at @neuripsconf.bsky.social to find out more!!
23.09.2025 08:57 — 👍 27 🔁 5 💬 1 📌 1
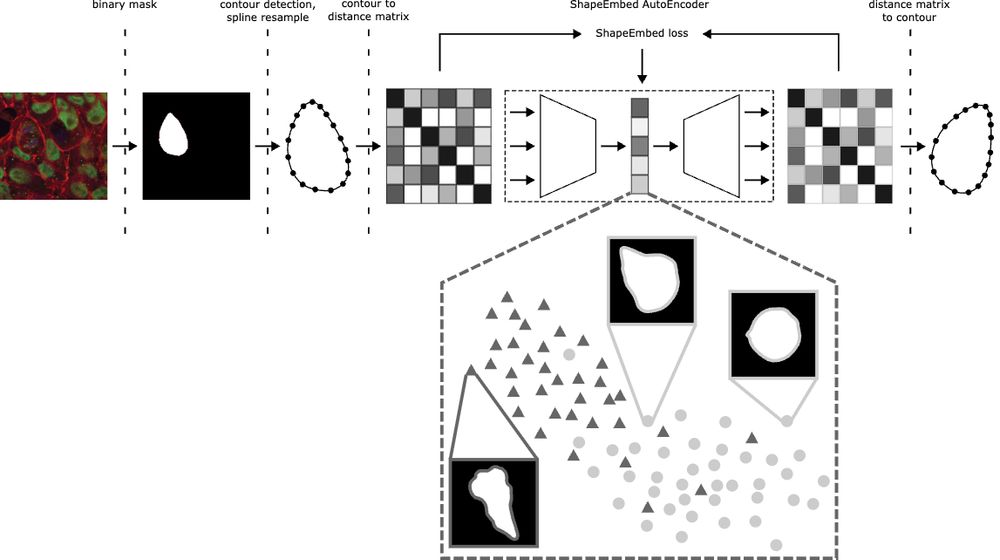
Happy to share that ShapeEmbed has been accepted at @neuripsconf.bsky.social 🎉 SE is self-supervised framework to encode 2D contours from microscopy & natural images into a latent representation invariant to translation, scaling, rotation, reflection & point indexing
📄 arxiv.org/pdf/2507.01009 (1/N)
23.09.2025 08:31 — 👍 71 🔁 26 💬 3 📌 5
(1/14) I’m happy and proud to introduce: SpinePy – a framework to detect the "spine" of gastruloids and measure biological and physical signals in a local dynamic 3D coordinate system. www.biorxiv.org/content/10.1...
12.09.2025 12:43 — 👍 63 🔁 24 💬 4 📌 2

Unified mass imaging maps the lipidome of vertebrate development
Nature Methods - uMAIA is an analytical framework designed to enable the construction of metabolic atlases at high resolution using mass spectrometry imaging data.
Happy to share our work on uMAIA, a framework for building metabolomic atlases from mass spectrometry imaging.
With uMAIA, we mapped the lipidome of zebrafish development, uncovering spatially organized metabolic programs .
www.nature.com/articles/s41...
#developmentalbiology #MSI #lipidtime
03.09.2025 10:40 — 👍 16 🔁 10 💬 2 📌 0
Cell tracking is never perfect, and it's important to understand the types of errors your solution contains.
Here is my stab at this: Divisualisations in @napari.org. github.com/bentaculum/d...
You spin tracks out upwards from the playing video. Green edges are correct, FP in magenta, FN in cyan.
30.07.2025 08:15 — 👍 53 🔁 17 💬 1 📌 0
Working in Martin’s group is an amazing experience, don’t hesitate to apply and reach out if you have any questions!
10.07.2025 20:39 — 👍 3 🔁 0 💬 0 📌 0
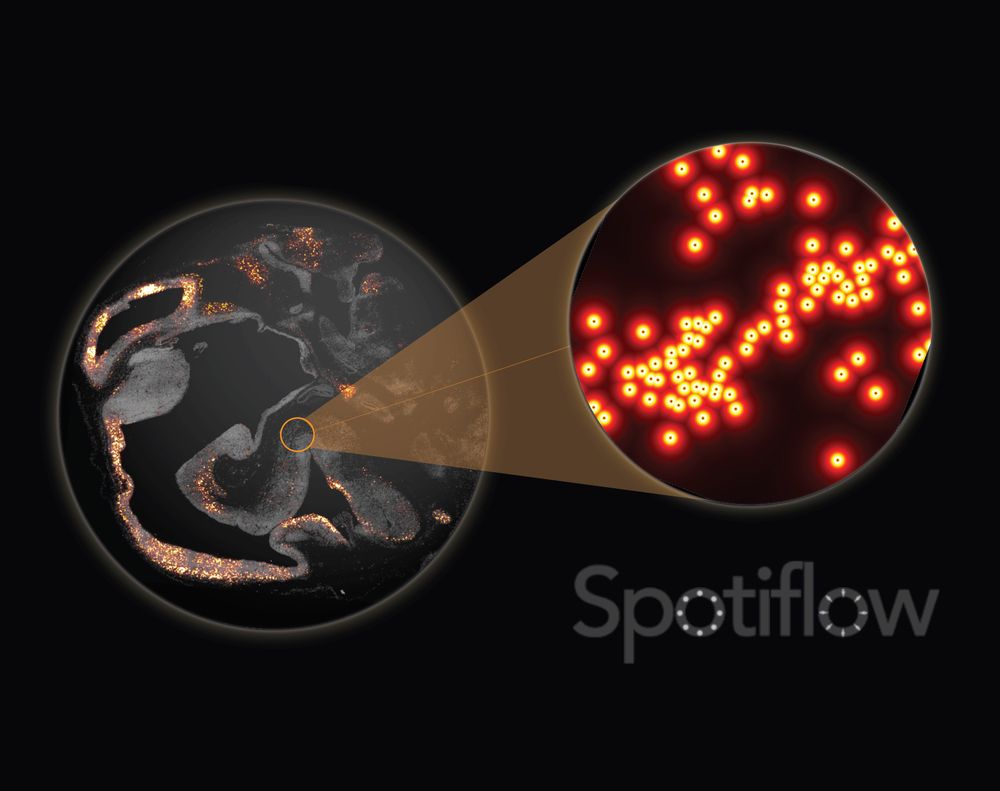
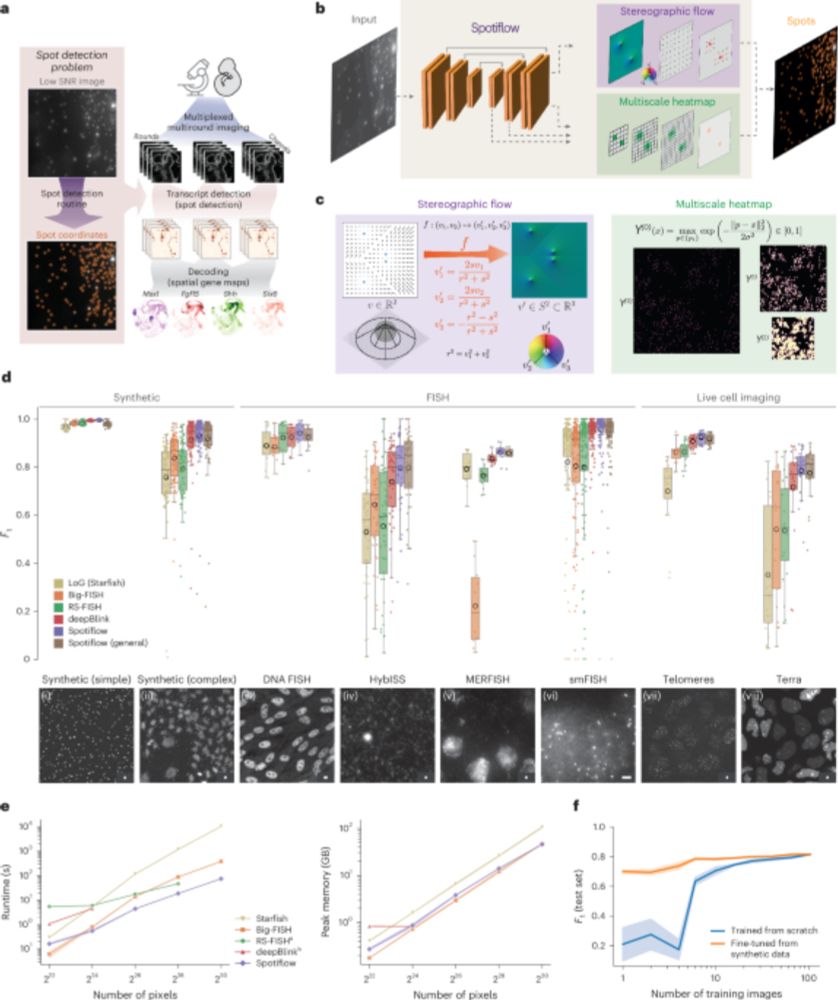
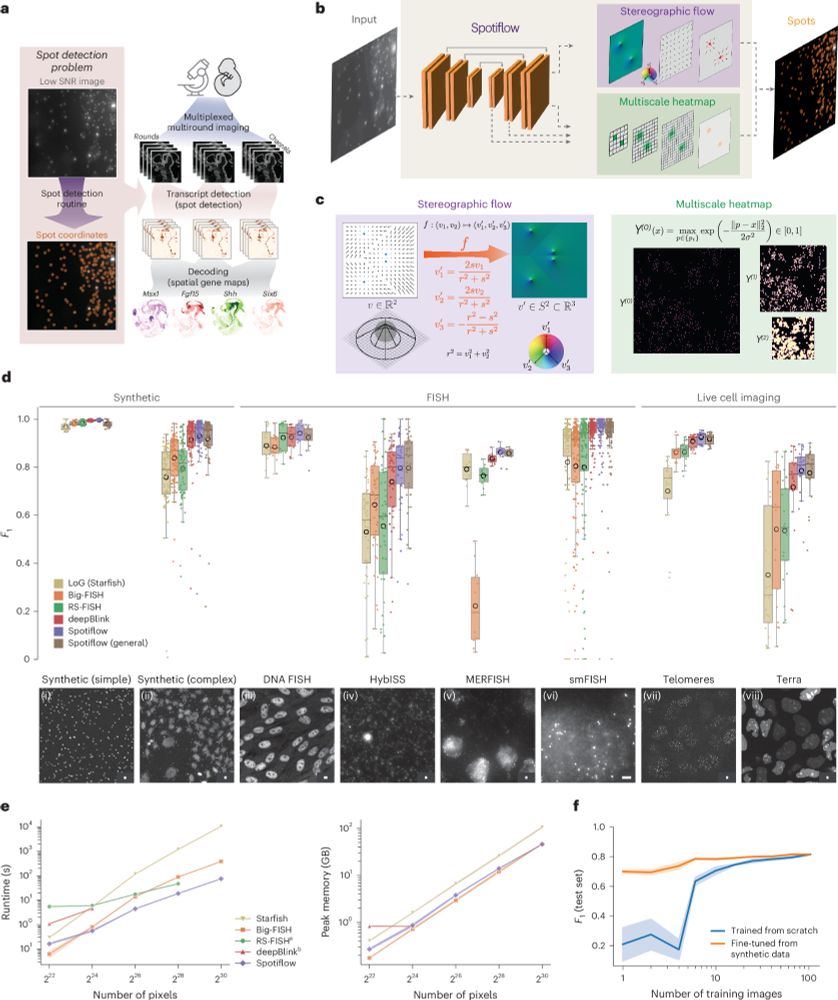
Out today in @natmethods.nature.com : Spotiflow, our transcript localization method for imaging-based spatial transcriptomics. Led by amazing PhD student @albertdm.bsky.social, joint work w @gioelelamanno.bsky.social at EPFL / @scadsai.bsky.social
www.nature.com/articles/s41...
rdcu.be/epIB7
06.06.2025 19:05 — 👍 109 🔁 37 💬 9 📌 2
A massive thanks to all the incredible people who contributed to this work, it's been an amazing ride! And a shoutout to all the people who have tried already tried Spotiflow and provided feedback, it's truly great to see the community using it :)
(N/N)
06.06.2025 18:58 — 👍 1 🔁 0 💬 0 📌 0

Results showing the effect of the number of training images on model performance for training from scratch and fine-tuning
We also offer several out-of-the-box pre-trained 2D/3D models. Fine-tuning a model on your own data is easy and accessible if you're not satisfied with the pre-trained ones, and we show Spotiflow is not particularly data hungry, so no need to spend a lot of time annotating!
(8/N)
06.06.2025 18:58 — 👍 0 🔁 0 💬 1 📌 0
We developed Spotiflow with the mindset of making it useful and usable, and as such it comes in different flavors: one can use it as a Python API, as a CLI or with a GUI using our @napari.org plugin! Hopefully more integrations will come soon ;)
(7/N)
06.06.2025 18:58 — 👍 4 🔁 3 💬 2 📌 0

Overview of EASI-FISH data + pairs of insets containing raw (left) and Spotiflow detections (right)
To be ready to handle today’s massive spatial biology data, we built a multi-GPU setup that e.g. processes a cloud-stored 159 GB EASI-FISH volume in under 10 minutes on 8×A100 GPUs. This can easily be deployed seamlessly on any modern compute cluster!
(6/N)
06.06.2025 18:58 — 👍 1 🔁 0 💬 1 📌 0
In a single molecule tracking experiment w/ Eftychia Kyriacou and Joachim Lingner from EPFL, Spotiflow allowed us to improve results by changing the input detections to Trackmate, showing that Spotiflow can be used as a drop-in replacement in existing workflows!
(5/N)
06.06.2025 18:58 — 👍 1 🔁 0 💬 1 📌 0
The context-awareness of Spotiflow allows for precise detection in challenging SNR conditions. This includes avoiding artifacts due to e.g. autofluorescence as we show with the beautiful P. dumerilii volumetric smFISH data from the Arendt and @ilastik-team.bsky.social teams @embl.org
(4/N)
06.06.2025 18:58 — 👍 0 🔁 0 💬 1 📌 0

Benchmarking results for different spot detection methods
Spotiflow achieves SOTA results on several datasets (iST, live imaging, DNA FISH) while being faster than commonly-used spot detection methods when used out-of-the-box on large-scale data, both in 2D and 3D!
(3/N)
06.06.2025 18:58 — 👍 0 🔁 0 💬 1 📌 0
Given an image or volume, Spotiflow uses a neural network to predict multiscale Gaussian heatmaps and the “stereographic flow” — a smooth embedding that maps the nD nearest-spot vector field to the (n+1)D sphere, avoiding issues at infinity and enabling precise localization
(2/N)
06.06.2025 18:58 — 👍 0 🔁 0 💬 1 📌 0

AI Method Improves Transcript Detection in Microscopy Images
Spotiflow improves large-scale transcript localization by combining deep learning with a novel geometric detection approach.
Get to know Spotiflow, an AI method that improves large-scale transcript localization by combining deep learning with a novel geometric detection approach.
Learn more:
👉 scads.ai/transcript-d...
Find the publication in Nature Methods:
👉 www.nature.com/articles/s41...
06.06.2025 10:14 — 👍 6 🔁 2 💬 0 📌 0
🔬🎤 As pre-announced a few days ago... please let us proudly present to you: 𝑴𝒊𝒄𝒓𝒐𝕊𝒑𝒍𝒊𝒕 - your ticket to imaging more, imaging more gentle, and/or imaging more efficient. 🔬
doi.org/10.1101/2025...
Like ❤️, repost 🔂, and most importantly... please send feedback ✉️ our way! 🙏
11.02.2025 18:02 — 👍 86 🔁 35 💬 2 📌 10
5'3' like DNA 🧬 bioimage analysis enthusiast & Cellular Immunology PhD –Graduate!– @ Dan Davis lab, Imperial. (she/her)
I'm a medical imaging and deep learning scientist at UCL. I love running, hiking and gaming
🔬🏃🏻♀️🥾🎮
PhD student at KCL.
Looking at muscle 💪🏼 with super-resolution fluorescence microscopy. 🔬
🍉🏳️🌈🏳️⚧️
Research Scientist in Loic Royer's team at Biohub | 🇧🇷
Ex-physicist in a biology institute, working on image analysis and building new hardware at the Francis Crick Institute
ORCID: 0000-0003-0994-5652
Scientist studying biological tissues with neuronal circuits inside. Tweets my own.
Aquí, tuitejo el què em passa pel cap molt de tant en tant.
trickymons@X
London-based neuroscientist 🧠 & research software engineer 💻 developing free and open-source tools for studying brains & behaviour, @neuroinformatics.dev @sainsburywellcome.bsky.social at UCL.
Committed to open, collaborative, and reproducible science.
Exploring RNA & RBPs in cancer: from cell edges to the chromatin. Microscopy lover, proteomics newbie. Likes computer sciences, physics & photography. Also an illustrator and graphic designer for Science and Biology (see: @STEMDORADOscimag)
Co-founder, core developer, and steering council member of @napari.org
Also other FOSSy and Science-y things.
I prefer Mastodon (profile at https://fosstodon.org/@jni) but begrudgingly checking things out here. 😂
BioImage Analysis Lead at AIC Janelia
The life and times of Steve the otter...
Postdoc in the Kleele lab, ETH Zürich | HFSP and EMBO fellow | Previously PhD in the Ewers lab, FU Berlin | mitochondria, neurons, super-resolution microscopy
group leader @ HHMIJanelia, #neuroscience + AI 🔬 #cellpose | diversity and open-science for better science | Ⓥ | she/her | https://mouseland.github.io
Still seeking M. Ulder for joint author paper.
Microscopes ✅ cats ✅ combining the two ❌
Lab --> https://www.kcl.ac.uk/research/culley-group
Microscopist & Python developer at Harvard Med. Creator @fpbase.org
BioImage Data Analysis at @crick.ac.uk: @crick-ia-team.bsky.social
Science → Reproducible as Data Analysis → Automated
Dubliner 🇮🇪 in London 🇬🇧 | Brunch sceptic | Trophy husband | Dissipator of masculine energy
🇲🇽🇯🇵
Computer Vision and Machine learning engineer at
CZ Biohub SF || Ph.D || views are my own || (he/him)
Post-doctoral researcher at BioVision Center, UZH || BioImage Data Scientist || Based in 📍Zurich || Born in Mallorca 🌊||
Bioimage analysis and computer vision enthusiast. Whenever I can, I do software too.
Ikerbasque | University of the Basque Country (UPV/EHU) | DIPC | Biofisika Institute
ORCID: 0000-0003-0229-5722
Argentinian physicist passionate about dissecting biology with microscopy.
PhD with P. Tomancak at MPI-CBG.
Postdoc with T. Dayton at EMBL-Barcelona.
She/her