I'm super excited to announce the first preprint of my PhD, together with Chenxi Ou and @sokrypton.org!
ML has revolutionized protein modeling, but crucial challenges remain. For example, we can't reliably predict complicated protein structures without MSAs, which limits what we can design.
10.12.2025 15:02 — 👍 31 🔁 6 💬 2 📌 1

eRMSF: A Python Package for Ensemble-Based RMSF Analysis of Biomolecular Systems
Understanding molecular flexibility and dynamics across different structural ensembles is essential for interpreting the behavior of complex biological systems. Here, we introduce eRMSF, a fast and user-friendly Python package built with MDAKit from MDAnalysis, designed to perform ensemble-based root mean square fluctuation (RMSF) analyses. Unlike traditional approaches limited to molecular dynamics trajectories, eRMSF extends flexibility analysis to ensembles generated by different methods, such as MD simulations, BioEmu (a deep learning tool for equilibrium ensemble prediction), subsampled AlphaFold2 (AlphaFold ensemble generation), and other computational or experimental sources. By enabling RMSF calculations across heterogeneous ensembles, eRMSF provides a unified framework to evaluate residue or atomic fluctuations in both simulated and predicted structures. Users can easily customize atom, residue, or region selections, tailoring analyses to specific research questions. This approach delivers high-resolution insights into localized motions, complements global stability assessments, and reveals dynamic regions often overlooked by single-method analyses. The repository for eRMSF is available at https://github.com/pablo-arantes/ermsfkit.
🎉 New publication in JCIM at @pubs.acs.org!
We present eRMSF, a Python package for ensemble-based RMSF analysis of biomolecular systems, supporting MD, PDB, and ML ensembles.
Proud to collaborate with Rodrigo Ligabue-Braun and @conradopedebos.bsky.social
pubs.acs.org/doi/10.1021/...
19.11.2025 11:08 — 👍 15 🔁 6 💬 2 📌 0
WOW!!! Fantastic news, Fiona! 🎉 Wishing you all the best, can’t wait to hear more about your new group!
12.10.2025 02:31 — 👍 0 🔁 0 💬 0 📌 0

🧠💥 ParametrizANI found a home!
Our paper is now published in JCIM!
Making ML-based force field parametrization open and easy for everyone.
With TorchANI, @rdkit.bsky.social & @openmm.org
🔗 pubs.acs.org/doi/abs/10.1...
@acs.org @giuliapalermo.bsky.social
09.10.2025 13:02 — 👍 3 🔁 0 💬 0 📌 1

Your protein moves, but when exactly? ⏱️
Meet eRMSF, our new Python tool that tracks fluctuations over time!
Traditional RMSF shows “how much.”
eRMSF shows “when.” 🔥
Preprint 🔗 doi.org/10.26434/che...
GitHub github.com/pablo-arante...
Try it on Colab → colab.research.google.com/github/pablo...
07.10.2025 13:12 — 👍 4 🔁 1 💬 0 📌 0
Exciting to see our protein binder design pipeline BindCraft published in its final form in @Nature ! This has been an amazing collaborative effort with Lennart, Christian, @sokrypton.org, Bruno and many other amazing lab members and collaborators.
www.nature.com/articles/s41...
27.08.2025 16:14 — 👍 305 🔁 109 💬 14 📌 11
Thank you, @olexandr.bsky.social! Yes, AIMNet2 is included in the notebook. Users can choose between several models: TorchANI, AIMNet2, and MACE-OFF. 😉
21.08.2025 10:54 — 👍 2 🔁 0 💬 0 📌 0
Thank you my friend! 🥰
20.08.2025 20:43 — 👍 0 🔁 0 💬 0 📌 0
When Pablo told me about this idea, I could see an immediate impact on drug discovery endeavors.
People can easily refine parameters of multiple dihedral torsion for hundreds of molecules and employ those in FEP simulations.
10/10 recommend 👍
20.08.2025 18:43 — 👍 4 🔁 1 💬 1 📌 0

GitHub - palermolab/ParametrizANI: ParametrizANI - Fast, Accurate and Free Dihedral Parametrization in the Cloud with TorchANI
ParametrizANI - Fast, Accurate and Free Dihedral Parametrization in the Cloud with TorchANI - palermolab/ParametrizANI
New from our lab!
🚀 ParametrizANI - a #NeuralNetworks tool for molecular parametrization.
Predicts potential energy surfaces with near-DFT/CC accuracy, at a fraction of the computational cost!
#AI #QuantumChemistry #CompChem #ML 🤖
chemrxiv.org/engage/chemr...
Try it:
github.com/palermolab/P...
20.08.2025 17:33 — 👍 37 🔁 9 💬 3 📌 0

a man with long hair and a beard is smiling and saying thank you .
ALT: a man with long hair and a beard is smiling and saying thank you .
Huge thanks to our co-authors Souvik Sinha & @giuliapalermo.bsky.social ! And deep gratitude to the @openmm.org, TorchANI team, Rotational Profiler developers, the "Making it Rain" team (@conradopedebos.bsky.social , @mdpoleto.bsky.social and Rodrigo Ligabue Braun) for their inspiration! 8/8
20.08.2025 13:34 — 👍 2 🔁 0 💬 0 📌 0

GitHub - palermolab/ParametrizANI: ParametrizANI - Fast, Accurate and Free Dihedral Parametrization in the Cloud with TorchANI
ParametrizANI - Fast, Accurate and Free Dihedral Parametrization in the Cloud with TorchANI - palermolab/ParametrizANI
Ready to try it? ✨ All our Colab notebooks are freely & publicly available on @github.com! Dive in to enhance your molecular studies. We're committed to fostering deeper insights & improved methodologies in the scientific community. Find ParametrizANI here: 7/8 github.com/palermolab/P...
20.08.2025 13:34 — 👍 0 🔁 0 💬 1 📌 0

a man with his mouth open and the words lightbulb moment written on the bottom
ALT: a man with his mouth open and the words lightbulb moment written on the bottom
A game-changer! ParametrizANI is perfect for drug discovery, helping evaluate candidates with high accuracy & speed. It's also an excellent resource for education, offering hands-on experience without complex setups, & allows professional customization. #DrugDiscovery 6/8
20.08.2025 13:34 — 👍 0 🔁 0 💬 1 📌 0

Our user-friendly Jupyter notebooks provide a complete workflow for dihedral parametrization, from SMILES strings to optimized force field parameters. We support both GAFF & @openforcefield.org force fields, ensuring compatibility for your simulations. #ForceFields 5/8
20.08.2025 13:34 — 👍 1 🔁 0 💬 2 📌 0

a cartoon character named spongebob is holding a box with the words " the cloud " written on it
ALT: a cartoon character named spongebob is holding a box with the words " the cloud " written on it
Accessibility is key! ParametrizANI runs on
@googlecolab.bsky.social, a "click-and-go" experience with free access to CPUs. No heavy parallel processing needed! We've parametrized molecules in less than 5 minutes on CPU. A big step for accurate, efficient small molecule parametrization! 4/8
20.08.2025 13:34 — 👍 0 🔁 0 💬 1 📌 0

a man says " my algorithm is insane " while wearing a baseball cap
ALT: a man says " my algorithm is insane " while wearing a baseball cap
A core component: our Python version of the Rotational Profiler code. This analytical algorithm efficiently computes classical torsional dihedral parameters by fitting empirical energy profiles to a reference curve. #ComputationalChemistry #Algorithms #ForceFields 3/8
20.08.2025 13:34 — 👍 0 🔁 0 💬 1 📌 0

a woman says yes that is painfully accurate in front of a crowd of people
ALT: a woman says yes that is painfully accurate in front of a crowd of people
How? ParametrizANI uses the robust PyTorch-based TorchANI program & ANI deep learning models (ANI-1x, ANI-2x). This predicts potential energy surfaces with near-DFT or coupled-cluster accuracy, at a fraction of the computational cost! #DeepLearning #TorchANI #AIforScience 2/8
20.08.2025 13:34 — 👍 1 🔁 0 💬 1 📌 0
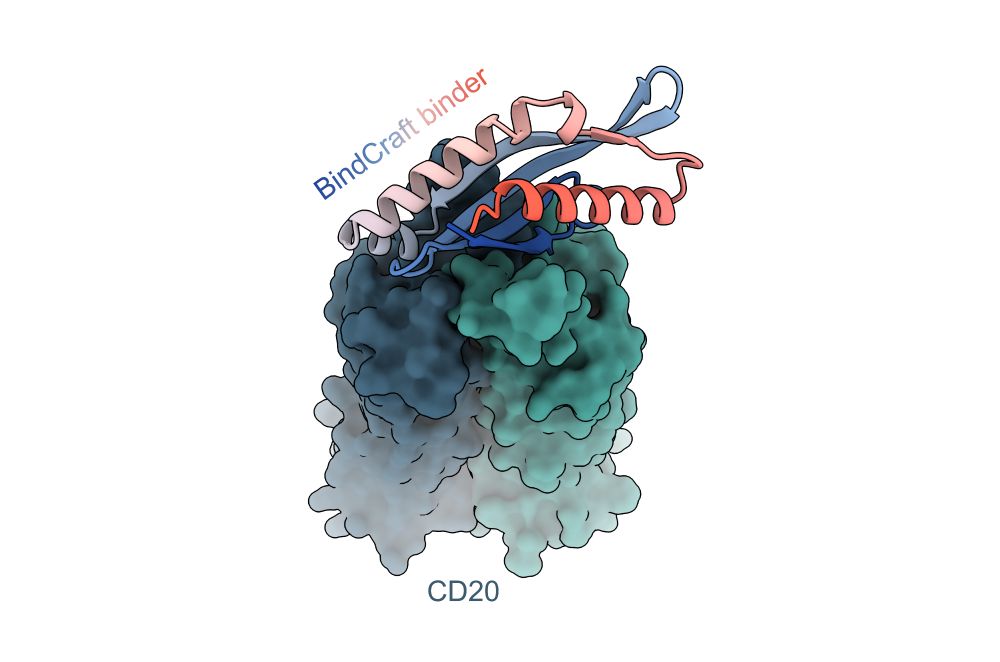
We have written up a tutorial on how to run BindCraft, how to prepare your input PDB, how to select hotspots, and various other tips and tricks to get the most out of binder design!
github.com/martinpacesa...
30.06.2025 19:45 — 👍 138 🔁 55 💬 4 📌 0

Front-page-style graphic titled “BREAKING NEWS” with photos of RFK Jr. and Dr. Bhattacharya in front of a government hearing chamber. Text reads: “NIH Scientists Sound the Alarm as Health Research Faces Historic Threat” and “NIH Employees Send Trump Cronies Scathing Wake-Up Call.”
🚨BREAKING: 300+ NIH employees call out the harm of censorship & politicized science in scathing email to Bhattacharya, demanding an end to political interference, a lift on funding freezes, & rehiring of fired staff whose work saves lives.
This is historic - insiders are blowing the whistle.
🧵(1/5)
09.06.2025 12:03 — 👍 4330 🔁 1401 💬 40 📌 73
Tem vaga ainda?
09.06.2025 01:17 — 👍 0 🔁 0 💬 0 📌 0
🚀 Excited to release BoltzDesign1!
✨ Now with LogMD-based trajectory visualization.
🔗 Demo: rcsb.ai/ff9c2b1ee8
Feedback & collabs welcome! 🙌
🔗: GitHub: github.com/yehlincho/Bo...
🔗: Colab: colab.research.google.com/github/yehli...
@sokrypton.org @martinpacesa.bsky.social
03.06.2025 01:30 — 👍 54 🔁 17 💬 1 📌 0

This is the kind of message that makes it all worth it.
When someone takes a moment to say thank you, it reminds me why we keep pushing forward—sharing tools, writing posts, and trying to make science more open and accessible. Grateful for the kind words!
08.04.2025 22:25 — 👍 12 🔁 3 💬 0 📌 0
Our latest paper just dropped in PNAS! 🎉
Turns out, CRISPR-associated transposons don’t just jump—they dance their way through DNA! 🕺🔬
Exciting times for genome engineering!
🧬 Read more in PNAS: www.pnas.org/doi/10.1073/...
#CRISPR #GeneEditing #PNAS #MDsimulations #CompChem
02.04.2025 19:12 — 👍 6 🔁 1 💬 0 📌 0
I have included side-chain reconstruction using HPacker in the BioEmu Notebook. Thank you to @martinsteinegger.bsky.social whose notebook provided inspiration for incorporating the cell to add side-chain reconstruction.
🔗 Try it on Google Colab: colab.research.google.com/github/pablo...
02.04.2025 00:34 — 👍 5 🔁 1 💬 0 📌 0
However, do you think it would be better to do 5000 sample compared to 1000 sample?
I ran some tests and compared an ensemble of 1000 samples with each containing 5000 frames. I did not observe any differences between them except for the number of frames.
24.03.2025 12:34 — 👍 1 🔁 0 💬 1 📌 0
Regarding your ideas, I loved the idea to compare an ensemble of WT and mutated protein using BioEmu, I'm working on a notebook to do that.
24.03.2025 12:34 — 👍 1 🔁 0 💬 2 📌 0
Hi Tareq, Thank you for all information regarding BioEmu. As I clearly described on my firt post, I did not develop the BioEmu, I just performed the implementation using Google Colab.
24.03.2025 12:34 — 👍 1 🔁 0 💬 2 📌 0
This code only supports sampling structures of monomers. You can try to sample multimers using two sequences you want to predict and connect them with a long linker, but in their experiments, this has not worked well.
21.03.2025 14:15 — 👍 2 🔁 0 💬 0 📌 0
PhD student @mit.edu, previously @deshawresearch.com, @harvard.edu
optimization, inverse problems, also proteins. ml at escalante. formerly: atomicai, xgenomes, broad, berkeley.
Ministra do Meio Ambiente e Mudança do Clima | Professora de história, senadora pelo Acre (1995-2011), deputada federal eleita por São Paulo.
Predoctoral Research Assistant at the Llorca lab @cniostopcancer.bsky.social working on mTORC1 signaling and cryo-EM ❄️🔬. Intrigued by the structural mechanisms of protein assemblies involved in endomembrane signaling and trafficking 🧪.
Postdoc in biophysics/biochemistry working on the interaction between the phosphorylated Tau protein and microtubules. Molecular dynamist. Tamer of IDRs and IDPs since 2022
(They/Them)
https://scholar.google.com/citations?user=4S1QUPgAAAAJ&hl=fr
Asst Prof in Chemical Engineering @VCU. Formerly postdoc @MITChemE. Machine learning and computational chemistry research.
mcgillresearchgroup.com
Ph.D. Student, Nobel laureate Carolyn Bertozzi and Longzhi Tan Labs, Stanford University🌲, Departments of Chemical and Systems Biology|Neurobiology|Sarafan ChEM-H, Passionate about Chemical Biology🧪, Neuroscience 🧠and Artificial Intelligence🤖
Theoretical & computational biophysics. Molecular information transfer in the cell. In-cell signaling, protein, membrane & lipid dynamics. Complex environments.
Lab @UIUC: pogorelov.scs.illinois.edu
PhD student @ #FUBerlin and MSCA fellow @ #TracktheTwin
// prev: physics @ #iisermohali.
https://jayashreenarayan.github.io/
Assistant prof at UCI School of Medicine | Molecular mechanisms of gene editors & Engineering precision editing tools to fix genetic diseases | CRISPR, structure, biochemistry
Computational structural biology and AI at the University of Maryland and Institute for Bioscience and Biotechnology Research. Decoding and engineering immune recognition.
Using quantitative, data-driven approaches to make synthetic chemistry more predictable. Phase II CCI #NSFFunded
🌐 https://ccas.nd.edu/
📷 https://www.instagram.com/nsf_ccas/
- Falo de games (foco em Nintendo) e outras coisinhas
- Ofertas Nintendo no ofertasnintendo.com.br
- E-mail ultranpodcast@gmail.com
- Podcaster no Ultra N Podcast youtube.com/ultranpodcast
PhD student@MIT
https://sites.google.com/view/yehlincho/home
Inherently disordered chemist @Technical University of Denmark. Sensors & functional materials. Pretty much winging it but having fun meanwhile. 🤙
Head of Partnerships and Development @consortium.lgbt | Chair @proudchangemakers.bsky.social | LGBTQ+ Columnist Exposed Magazine
#LGBWithTheT Proud Queer 🏳️🌈🏳️⚧️
www.heatherpaterson.co.uk
Retired physics prof, forever lefty political nerd. Born at 315 ppm. In a relationship. Reform the Democratic Party! NO NO NO on the SAVE Act!
Reconstitute ICE - you can't make good ICE from polluted water!
















