MIT Technology Review article about our research on human embryo models in this day & age.
I didn't agree to interview because what i find as over-sensational writing style & i disagree with several things..
BUT Antonio managed to put together an interesting mix of events and ideas worth reading.
25.10.2025 21:05 — 👍 16 🔁 4 💬 0 📌 0
Last day of intense science discussions and presentations at the HFSP Science Summit Japan was held in Kyoto university. It has been an amazing 5 days of listening to great scientists from Japan and all over the world. An unforgettable visit.
04.10.2025 12:15 — 👍 4 🔁 0 💬 0 📌 0
Another great day in Japan science summit with Intriguing science and science policy discussions at University of Tokyo.
03.10.2025 06:50 — 👍 4 🔁 0 💬 0 📌 0
This was a real fun event!
01.10.2025 09:33 — 👍 2 🔁 0 💬 0 📌 0

EEE meeting is BACK! Early Embryogenesis & Epigenetics conference in Berlin 02/2026.
Checkout great program and over 12 slots for (not so) short talks for submitted abstracts!. Early registration now open -
w.molgen.mpg.de/embryo2026
28.09.2025 00:00 — 👍 29 🔁 12 💬 0 📌 1

Having fun @amartinezarias.bsky.social party! 🙏🏻🎉💐 Great meeting #InHoxWeSortOfTrust
19.09.2025 19:59 — 👍 33 🔁 5 💬 0 📌 2

HFSP Science Summit Japan 2025
🧪 HFSP & partners present Science Summit Japan 2025!
🗓️Oct 1–4 📍Tohoku University, RIKEN, UThe University of Tokyo & Kyoto University.
✨ Nobel Laureates, HFSP Nakasone Awardees & top researchers engaging with young scientists.
More info 👉https://bit.ly/45WGRWM
03.09.2025 09:40 — 👍 5 🔁 1 💬 0 📌 0

Awards + Nominations — International Society for Stem Cell Research
The ISSCR Awards portfolio recognizes outstanding achievements and research potential in the field of stem cell research around the world. The ISSCR awards honor scientists’ innovative work to harness...
Shout out re @isscr.org awards nomination deadline SEP 30 2025! Please note the great revision & efforts that have been made to recognize excellence in industry, policy makers, clinical translation etc & from across the globe. Self-nominations are equally welcomed!
www.isscr.org/awards
28.08.2025 11:50 — 👍 2 🔁 2 💬 0 📌 0
Super congrats dear Richard
27.08.2025 19:38 — 👍 0 🔁 0 💬 1 📌 0
Tnx to @hfspo.bsky.social for humbling opportunity to present our work #HFSPNakasoneAward lecture.
Tnx to Hanna lab, current & former members, collaborators, mentors. @weizmanninstitute.bsky.social for critical backing, @isscr.org for guidelines, funding agencies & dear philanthropists for funding
14.08.2025 11:59 — 👍 11 🔁 0 💬 0 📌 0
thank you dear luca for organizing this great meeting and having me. i had great time attending the fantastic diverse sessions and talks. I look forward for next one me and my lab embers would certainly become regular attendees.
14.08.2025 11:54 — 👍 1 🔁 0 💬 0 📌 0

12/n last one (plenty more but u should get the point by now) - iEFCs=60h in Preprint are shown to have a mix of PrE/Epi/TE , while in Paper they are homogeneously labeled in Magenta?
12.08.2025 06:45 — 👍 0 🔁 0 💬 0 📌 0

11/n in relation to the above concern, another panel of Paper in Fig.3F 32C ICM are marked (left) and according to this analysis on dataset used they also highly overlap with 16-Cell labeled in green? yet again why drown in scRNA-seq when immunostainings r so easy to solve this ?
12.08.2025 06:44 — 👍 0 🔁 0 💬 1 📌 0

10/n some meta-analysis was done to claim iEFC fall on 8 cell. Upon close inspection - 8C and 32ICM r coloured similarly! and there is NO arrow pointing to 32ICM on the UMAP. it seems 32ICM cells are nicely overlapping with iEFCs? where is 64cell ICM? But again, simple scRNA-seq above says it all!
12.08.2025 06:44 — 👍 0 🔁 0 💬 1 📌 0

9/n Straightforward scRNA-seq analysis of 60h=iEFC in Preprint also showed cells are not going backwards in development, but forward. Gata6+ cells express Sox17 and Pdgfra which are not 8-16cell markers but of good old E4.5 PrE cells.
12.08.2025 06:43 — 👍 0 🔁 0 💬 1 📌 0

8/n Strangely straightforward scRNA-seq analysis of 60h=iEFC in Preprint showing rare CDX2/GATA6 coexpression & and that iEFC is basically a mix of Epi/PrE/TE, was NOT included in the published Paper although same dataset was used in both. & Nanog is reduced in TE-&PrE-like cells.
12.08.2025 06:42 — 👍 0 🔁 0 💬 1 📌 0

7/n Preprint: 3/3 iEFC stainings show no/rare CDX2+/GATA6+ (include the same 2 above) and one in Preprint Fig1C, which was replaced in Paper Fig 1E with one that shows fuzzy coexpression. In sum 3 out of 4 immunostainings shown, rare CDX2/GATA6 co-expression!
12.08.2025 06:42 — 👍 0 🔁 0 💬 1 📌 0

6/n why drown in intracellular stainings? Immunostaining for these markers r very easy to do in EFC=60h (names of same between Paper & Preprint tinyurl.com/tu6cv3cd). No quantitative & wideview stainings r shown. 2/3 iEFC stainings in Paper show no/rare CDX2+/GATA6+.
12.08.2025 06:42 — 👍 0 🔁 0 💬 1 📌 0
5/n why is this important? We need to distinguish btwn: 1)Naive ESC reverting back to 8-16 cell "EFCs"= most cells co-expressing OCT4/CDX2/GATA6 2)Naive ESC partially/on their way differentiating into TE or PrE = resulting in OCT4+ cells with CDX2+ or GATA6+, NOT CDX2/GATA6 co-expressed
12.08.2025 06:41 — 👍 0 🔁 0 💬 1 📌 0
4/n These discrepancies using same data controls & samples between preprint and paper are very concerning. Gating strategy & methods not shown explained in paper, but this change shows the authors are determining gating based on signal detected in ESC sample and not EFC (60h) own negative controls.
12.08.2025 05:53 — 👍 0 🔁 0 💬 1 📌 0

3/n Intracellular FACS staining for TFs is a "dirty assay", each sample must have its OWN unstained and ISOTYPE MATCHED CONTROL to ensure true gating 4 positive staining. Wasn't done! & Preprint had GATING showing low%, was CHANGED in the Paper resulting in boost up to >85%!
12.08.2025 05:52 — 👍 0 🔁 0 💬 1 📌 0

2/n 2/n Li et al. purported a concept of transiently capturing EFCs based on coexpression of OCT4/CDX2/GATA6 in 8-16cell embryos, after 60h of chemical treatment. Main proof shown is based on triple+ FACS intracellular staining for these genes (& NOT using triple reporter ESC)
12.08.2025 05:51 — 👍 1 🔁 0 💬 1 📌 0
🤩 Very excited to share our new work! We have derived euploid and aneuploid trophoblast organoids and extra-embryonic mesoderm cell lines from early human embryos. In doing so, we have characterised the tissue requirements for their specification. If you want to know more, continue reading….
09.08.2025 08:03 — 👍 37 🔁 11 💬 1 📌 0

end / Thanks to all Hanna lab members, great collaborators, funders and our lead Heroes from Turkey,
Gulben and Alperen (BSKY less) for the hard work despite of all the noise around.
07.08.2025 18:23 — 👍 3 🔁 0 💬 0 📌 0

15/n But rather, naive PSCs have minimal plasticity in 2i/LIF shown by Brickman tinyurl.com/yhzrducz & our conditions boost amplify this propensity by using alternative signaling conditions. Naive PSCs are de facto totipotent and have "multiple personalities!" . "The Matrix" continues to inspire :)
07.08.2025 18:16 — 👍 3 🔁 0 💬 1 📌 0

14/n In summary, embryo founder-like cells r neither induced nor involved in TF-SEM formation. there is no evidence that mouse nPSCs need to first go to an earlier "embryo founder cells" & only then give rise to TE/PrE/Epi.
07.08.2025 18:14 — 👍 0 🔁 0 💬 1 📌 0
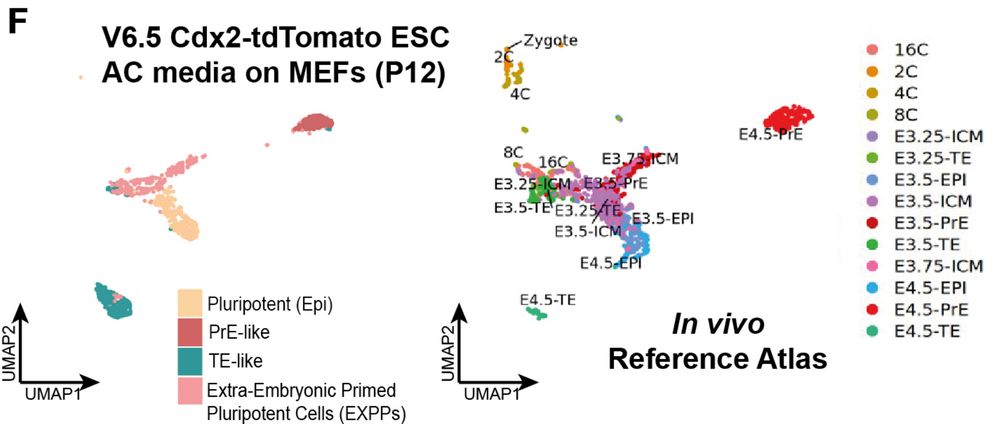
13/n AC-MEF cells were blastoid competent! and TF-SEM competent. They also lacked any 2C-like or 8-16 cell-morula-like embryo founder cells during expansion!
07.08.2025 18:14 — 👍 1 🔁 0 💬 1 📌 0

12/n Naive ESCs in AC-MEF conditions generated advanced E8.5 TF-SEMs. So we developed a rapid induction regimen , and also along-term induction and maintenance based route to make mouse TF-SEMs.
07.08.2025 18:13 — 👍 1 🔁 0 💬 1 📌 0
PhD candidate in Aktas Lab @MPI for Molecular Genetics in Berlin. 🧬🔬
Passionate about Evolution and Molecular Biology.
PhD Student @EMBL | Marine microbiology & ecotoxicology🌊🧪| Vincent group @floravincent 🏋️♀️| obsessed with sustainability🌿 (she/her)
Professor of Biophysics -
@ISTAustria Using physics to understand biological questions, such as how embryos grow!
Antifa, Science, Sustainability & Swing. Head of Scientific Coordination at the Max Planck Institute for Molecular Genetics, Berlin. Opinions are my own.
The Max Planck Institute for Molecular Genetics in Berlin. We want to understand how the genome works and how it is regulated, especially during embryonic development & disease. Imprint: https://www.molgen.mpg.de/impressum
EMBO/MSCA postdoctoral fellow at Francis Crick Institute in the Briscoe lab. 🇬🇧
Previously PhD Stainier Lab (heart dev 💓) @MPI-HLR 🇩🇪
Developmental biologist, lineage trajectories, cell fate decisions, cellular interactions, organ formation.
Postdoc - Duboule Lab - @college-de-france.fr @cirbcdf.bsky.social
Formerly PhD student - Furlong Lab - @EMBL.org
Interested in gene expression and genome topology regulation during embryogenesis
From 🇮🇹 , then Heidelberg 🇩🇪, now Paris 🇫🇷
Cell Stem Cell publishes research and reviews at the forefront of stem cell biology, bioengineering, and regenerative medicine. Posts by Cell Stem Cell editors.
PI @ CIID, Heidelberg, and @DZIF. Mom of two and scientist in love with nuclear architecture - NPC- RNA processing, metabolic switches in infected immune blood and brain cells 🧬🔬🧬🦠🧫
Postdoc at @Duboule lab. Biologist interested in mechanisms of gene regulation and chromatin 3D architecture
PI @ Gladstone Institutes & UCSF. Molecular technologies & the genomics / molecular biology / biochemistry of gene regulation. Views here mine & do not represent those of my affiliated institutions.
Scientist at IMP in Vienna. Excited about gene expression regulation and its encoding in our genomes - enhancers, transcription factors, co-factors, silencers, AI.
🔬 Assistant Prof, Pathology @Duke | Director, Clin Micro Lab
🧫 Former Clin Micro Fellow @Memorial Sloan Kettering
👩🏻🔬 Former Postdoc @broadinstitute.org
🎓 PhD @The Rockefeller University
Focus: Diagnostics, AMR, Structural Biology
I enjoy playing with family, reading science fiction and history, gardening, board games, and jazzercise. I'm a father, husband, Navy veteran, and graduate student studying computer science at George Mason University.
Mastodon: https://scicomm.xyz/@David
Genomics, Machine Learning, Statistics, Big Data and Football (Soccer, GGMU)
Post-doc at the Morris lab @BWH @harvardmed.bsky.social. Previously PhD at the Gottgens lab @scicambridge.bsky.social. Interested in developmental biology, single-cell biology, bio-informatics.
Professor of #StructuralBiology @karolinskainst.bsky.social studying #egg-#sperm interaction at #fertilization & #ZonaPellucida (ZP) module proteins
Member of #EMBO & the #NobelAssembly at Karolinska Institutet
More @ http://jovinelab.org
Else Kröner Fresenius Professor & Chair of Translational Nutritional Medicine @tum.de | PI @helmholtzmunich.bsky.social | Former @lmumuenchen.bsky.social @harvard.edu @uni-hamburg.de | Banner by @albamena.bsky.social
Homepage: https://www.mls.ls.tum.de/tnm



















