Congratulations, Arnaud!
21.08.2025 18:48 — 👍 1 🔁 0 💬 0 📌 0
This is not a HiC map! Ever wondered if multiple enhancers get activated simultaneously? We measured chromatin accessibility on thousands of molecules by nanopore to create genome-wide co-accessibility maps. Proud of @mathias-boulanger.bsky.social @kasitc.bsky.social Biology in the thread👇
18.08.2025 13:00 — 👍 113 🔁 34 💬 10 📌 0

Activity of most genes is controlled by multiple enhancers, but is there activation coordinated? We leveraged Nanopore to identify a specific set of elements that are simultaneously accessible on the same DNA molecules and are coordinated in their activation. www.biorxiv.org/content/10.1...
18.08.2025 12:23 — 👍 98 🔁 39 💬 2 📌 2
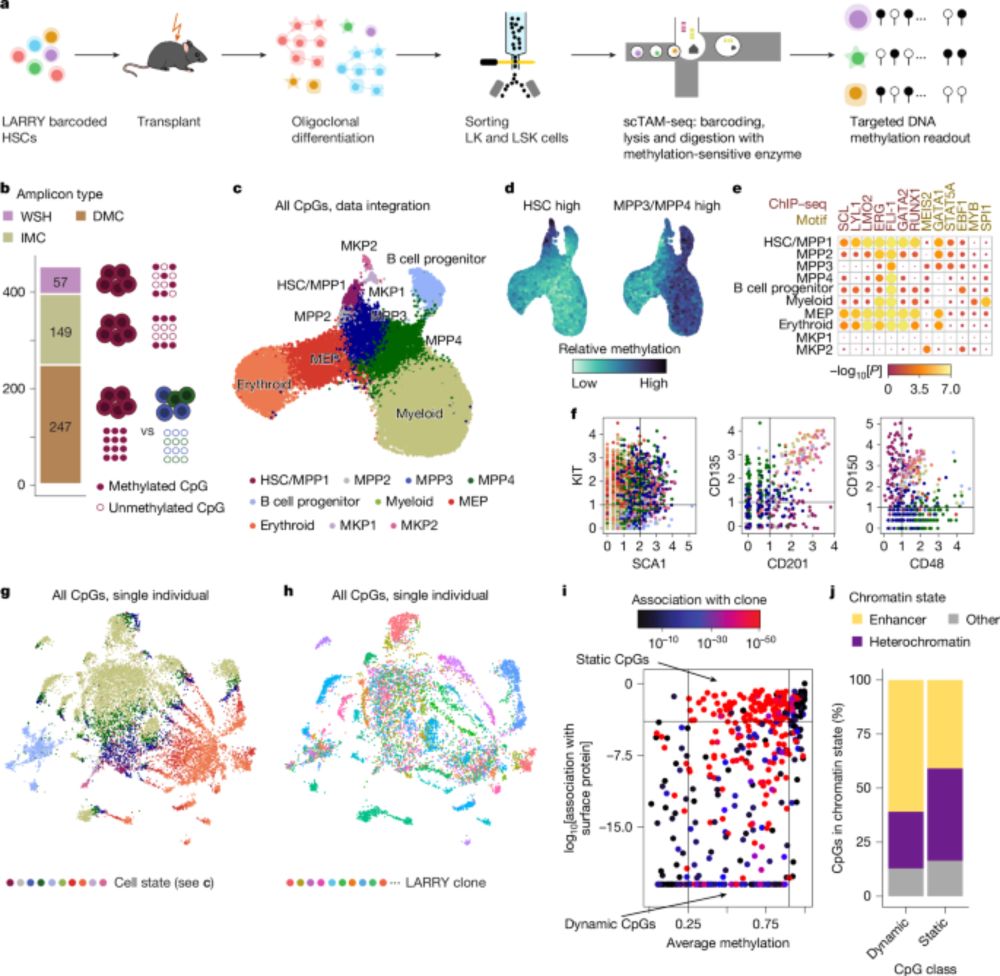
Excited to see this published with additional data following our preprint a while back. Cool combination (in our biased view) of controlled TF expression and machine learning to decode chromatin sensitivity. www.sciencedirect.com/science/arti....
07.08.2025 16:20 — 👍 114 🔁 49 💬 2 📌 3
Now available in its final form @narjournal.bsky.social !
doi.org/10.1093/nar/...
Find out how we can reconstruct enhancer activity in vivo in the Drosophila embryo using scRNAseq data and Optimal Transport.
23.07.2025 07:44 — 👍 23 🔁 7 💬 3 📌 1

#EMBLsinglemolecule was a blast! Happy to host the birth of single molecule genomics as a field and to think about the future with the microscopy crowd. Thanks to participants and organizers!
18.07.2025 12:04 — 👍 48 🔁 5 💬 1 📌 2
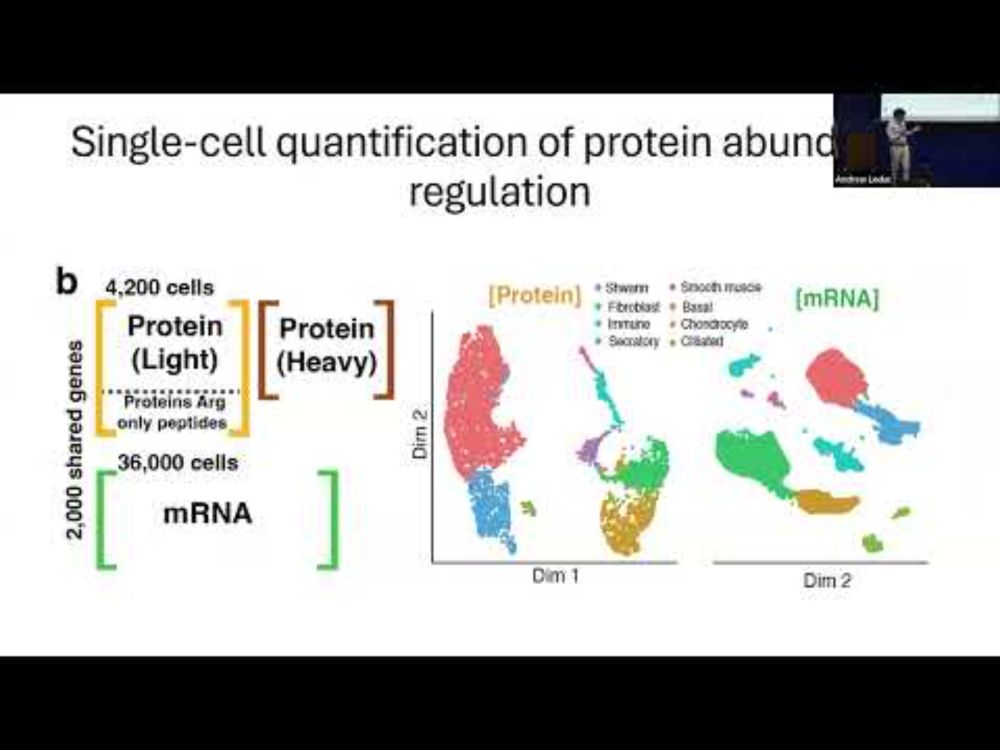
YouTube video by Nikolai Slavov
Quantification of gene expression control in a mammalian tissue at single cell resolution | SCP2025
The talk by @andrewleduc.bsky.social at #SCP2025 is on YouTube:
𝐐𝐮𝐚𝐧𝐭𝐢𝐟𝐢𝐜𝐚𝐭𝐢𝐨𝐧 𝐨𝐟 𝐠𝐞𝐧𝐞 𝐞𝐱𝐩𝐫𝐞𝐬𝐬𝐢𝐨𝐧 𝐜𝐨𝐧𝐭𝐫𝐨𝐥 𝐢𝐧 𝐚 𝐦𝐚𝐦𝐦𝐚𝐥𝐢𝐚𝐧 𝐭𝐢𝐬𝐬𝐮𝐞 𝐚𝐭 𝐬𝐢𝐧𝐠𝐥𝐞 𝐜𝐞𝐥𝐥 𝐫𝐞𝐬𝐨𝐥𝐮𝐭𝐢𝐨𝐧
youtu.be/adkY6txDyqs?...
11.06.2025 12:29 — 👍 9 🔁 3 💬 0 📌 3
🚨 Only a few more days to REGISTER for the TriRhena Gene Regulation Club in Freiburg.
Whether you are new or an established researcher in the Basel-Freiburg-Strasbourg region, this is your meeting when working in transcription, chromatin & gene regulation!
✍️ www.ie-freiburg.mpg.de/gene-regulat...
03.06.2025 12:59 — 👍 4 🔁 4 💬 0 📌 1
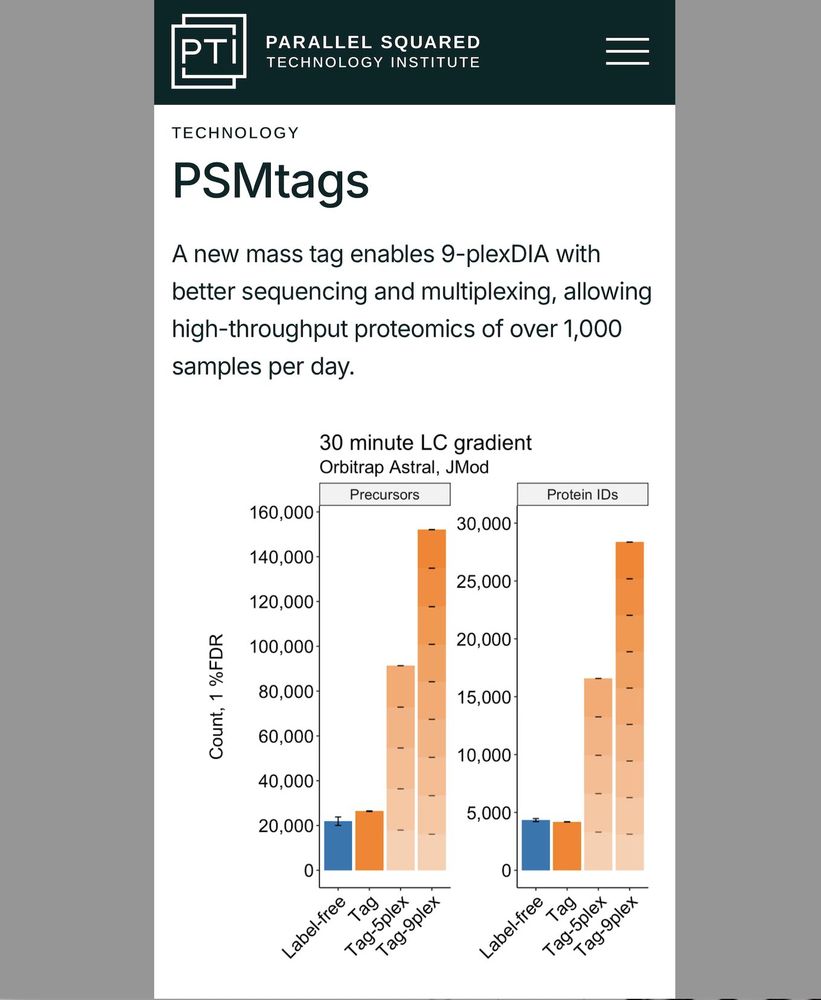
🟦 Label-free single-cell proteomics (blue bars).
🟧 Multiplexed single-cell proteomics affords higher throughput (orange bars).
⬛️ Proteome depth & quantitative accuracy are comparable.
We aim to make the 🟧 bars taller.
www.parallelsq.org/psmtags
www.biorxiv.org/content/10.1...
31.05.2025 13:11 — 👍 13 🔁 7 💬 0 📌 0
For Spanish speakers, this is a fantastic podcast about science and much more!
29.05.2025 17:26 — 👍 1 🔁 1 💬 1 📌 0
No doubt, this is one of the most exciting meetings of the year! And we will present our new method, HiddenFoot 😉: www.biorxiv.org/content/10.1...
28.05.2025 21:45 — 👍 6 🔁 2 💬 0 📌 0

‼️Last chance to join us in person for #EMBLSingleMolecule! Attend talks, network, socialise, and experience the atmosphere on the campus 🤩
🎫 Register by 3 Jun 👉🏼 http://s.embl.org/grg25-01-bl
🧬RNA processing
🧬Translation
🧬Transcription & chromatin regulation
🧬Method development
🧬Theory
27.05.2025 13:45 — 👍 5 🔁 4 💬 0 📌 2
Very cool, Lars! Congrats!
22.05.2025 06:25 — 👍 4 🔁 0 💬 0 📌 0
Just in case you missed this and it’s something that might interest you. #SingleMoleculeBiology, #Chromatin, #Transcription, #TF, #Nucleosome, #PolII, #Biophysics, #MachineLearning, #ComputationalBiology 👇
21.05.2025 20:31 — 👍 6 🔁 0 💬 0 📌 0
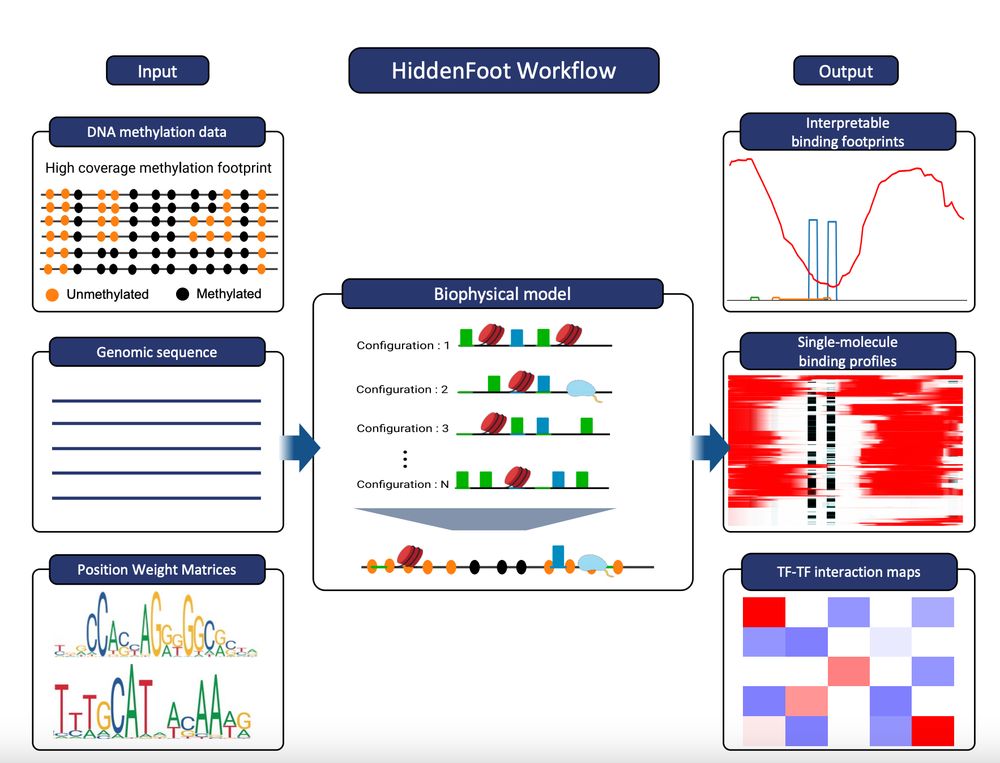
Time for a short thread! We developed HiddenFoot, a biophysics-inspired approach to decode single-molecule footprinting data and infer TF, nucleosome, and RNA Pol II binding profiles on individual DNA molecules. One molecule at a time! www.biorxiv.org/content/10.1...
1/6
19.05.2025 18:55 — 👍 54 🔁 18 💬 2 📌 2
Thanks!
20.05.2025 04:31 — 👍 2 🔁 0 💬 0 📌 0
Thanks!
19.05.2025 21:48 — 👍 1 🔁 0 💬 0 📌 0
I'm neighboring the chromatin biology field at the moment and there is a lot I need to understand. Especially, the specific techniques/methodologies in use to study chromatin, however this one looks 🔥🔥🔥.
Many congratulations 👏🏻 🎉 to all those involved!
Hey #ChromatinSky people where are you at 🤩?
19.05.2025 21:44 — 👍 6 🔁 1 💬 1 📌 0
Muchas gracias!!!
19.05.2025 19:40 — 👍 1 🔁 0 💬 1 📌 0

Finally, if you've made it this far, you might be interested in testing the code. Feedback is very welcome! github.com/MolinaLab-IG...
6/6
19.05.2025 19:29 — 👍 4 🔁 0 💬 1 📌 0

HiddenFoot also works with Fiber-seq data! It reveals chromatin structure and nucleosome occupancy heterogeneity at single-molecule resolution driven by TF binding.
5/6
19.05.2025 19:24 — 👍 3 🔁 0 💬 1 📌 0

HiddenFoot resolves Pol II and nucleosome occupancy at the HIV-1 promoter, molecule by molecule. Under transcriptional inhibition (TLD), Pol II footprints vanish, while nucleosome occupancy at the +1 and −1 positions increases.
4/6
19.05.2025 19:19 — 👍 0 🔁 0 💬 1 📌 0

HiddenFoot infers pairwise interaction energies from single-molecule data and compares them to simulated equilibrium profiles to distinguish true TF–TF cooperativity from nucleosome-mediated co-binding.
3/6
19.05.2025 19:18 — 👍 0 🔁 0 💬 1 📌 0

HiddenFoot is a thermodynamics-based model that integrates known TF PWMs and nucleosome occupancy to efficiently evaluate all possible non-overlapping binding configurations. It fits model parameters using both stochastic gradient descent and MCMC.
2/6
19.05.2025 19:07 — 👍 1 🔁 0 💬 1 📌 0

Time for a short thread! We developed HiddenFoot, a biophysics-inspired approach to decode single-molecule footprinting data and infer TF, nucleosome, and RNA Pol II binding profiles on individual DNA molecules. One molecule at a time! www.biorxiv.org/content/10.1...
1/6
19.05.2025 18:55 — 👍 54 🔁 18 💬 2 📌 2
Predoctoral Research Assistant at the Llorca lab @cniostopcancer.bsky.social working on mTORC1 signaling and cryo-EM ❄️🔬. Intrigued by the structural mechanisms of protein assemblies involved in endomembrane signaling and trafficking 🧪.
JupyterLab is my Winamp visualizer.
Leading the Computational Systems Medicine Lab @uconnhealth.bsky.social | Mathematician exploring life’s equations and algorithms | Tri-National | Cancer Survivor | Lifelong Learner
Senior Bioinformatician / data analyst at @EMBL GeneCore - Ph.D in health biology and genomics | Former postdoc at @EMBL Krebs lab - PhD student at @igmm
Biophysicist, Group Leader at MPIPZ. Studying the multicellular dynamics in plants, combining theory and experiments. Also, fascinated by music and dance.
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
CNB-CSIC researcher, head of the Cell Senescence, Cancer and Aging Lab (ColladoLab) at CiMUS-USC, IDIS, Santiago de Compostela, Spain.
SNF Ambizione Fellow at the FMI in Basel, Switzerland. Studying how dynamic chromatin regulates development. https://methstep.github.io/
Doctoral candidate in the Cissé lab @mpi-ie.bsky.social | Combining experiment and computation | Passion for the mountains and music
scientist - epigenetics, genomics, synthetic biology
Scientist & Communicator. Fascinated by cells across scales - 🌎, 🔬and ⏳. PI @GIMM, EMBO member. Former Director @IGC, PD @Cambridge University, PhD @UCL/IGC. My motto: "Science from all for all". Mom of 2 wonderful girls. Views are my own.
Research, news, and commentary from Nature, the international science journal. For daily science news, get Nature Briefing: https://go.nature.com/get-Nature-Briefing
Group leader @crg.eu | blood, single cell, synthetic genomics | Dad | 🇩🇪🇪🇸🇹🇿 | 🚵🧗🏃
Inserm researcher and group leader at Institut Curie. Linking cancer immunity, immunotherapy, transcription, and AI/ML. https://institut-curie.org/team/waterfall
(she/her) Computational biologist and post-doc scientist in the Greenleaf and Kundaje labs at Stanford. Interested in understanding how cells know what to become (transcription factors, gene regulation, dev bio, open science) www.selinjessa.com
Scientist in North Carolina studying chromatin remodelers and transcription factors. @fnucleosome.bsky.social coorganizer
ORCID: 0000-0002-6256-144X
Views and posts are all my own
Uncovering the beauty of embryonic development with functional RNA biology tools. | TT Prof at @ Carl Zeiss Center for Synthetic Genomics & KIT | http://modiclab.org/
Postdoctoral Fellow, Genome Biology Unit, EMBL Heidelberg
Genome structure and function at the Technion Faculty of Medicine