New Tuesday (Oct 28) in Heidelberg and online: kick off event of the Center for Synthetic Genomics: program and link to zoom: www.syn-gen.de/kick-off
24.10.2025 11:19 — 👍 4 🔁 5 💬 0 📌 1New Tuesday (Oct 28) in Heidelberg and online: kick off event of the Center for Synthetic Genomics: program and link to zoom: www.syn-gen.de/kick-off
24.10.2025 11:19 — 👍 4 🔁 5 💬 0 📌 1Beautiful mechanistic story, how QKI by bridging intronic motifs with U6 snRNA ensures inclusion of weak 5’ splice sites that are essential for building sarcomeres. Congrats @kurianlab.bsky.social and @evannostrandlab.bsky.social labs & all involved.
08.09.2025 15:24 — 👍 3 🔁 1 💬 2 📌 0
New paper from the Trcek lab, with Siran Tian as the lead author - congratulations Siran!: www.nature.com/articles/s41...
30.08.2025 23:13 — 👍 18 🔁 8 💬 1 📌 2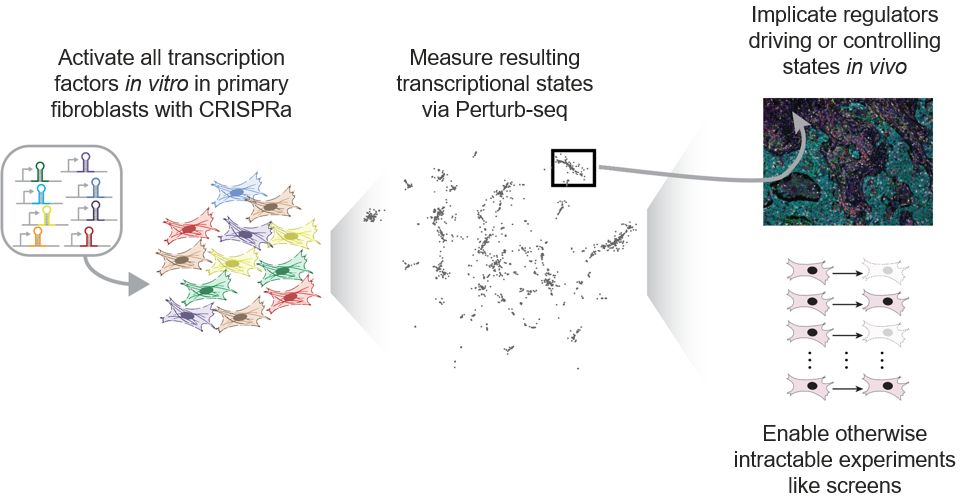
Our paper is now out in final form at Nature Genetics! For those who missed the preprint, we used large-scale Perturb-seq targeting transcription factors to push primary fibroblasts into diverse transcriptional states, including those observed in cell atlas studies.
06.08.2025 15:14 — 👍 74 🔁 29 💬 3 📌 1

Explore RNA's role in epigenetics, DNA repair, transcription & genome organisation - directly with leaders from academia & industry at #GenReg26!
Remaining talks to be filled from submissions - register & submit by 04 Sep 2025 to join us in Mexico.
Speakers, details & registration➡️ bit.ly/46JQbze
I’m a bit late to the game here, but wanted to highlight this very clever technique from @paraspeckle.bsky.social’s group! Congrats to all authors!!
02.08.2025 18:55 — 👍 7 🔁 2 💬 0 📌 0
A day late but STILL IMPORTANT
02.08.2025 16:14 — 👍 31 🔁 7 💬 2 📌 1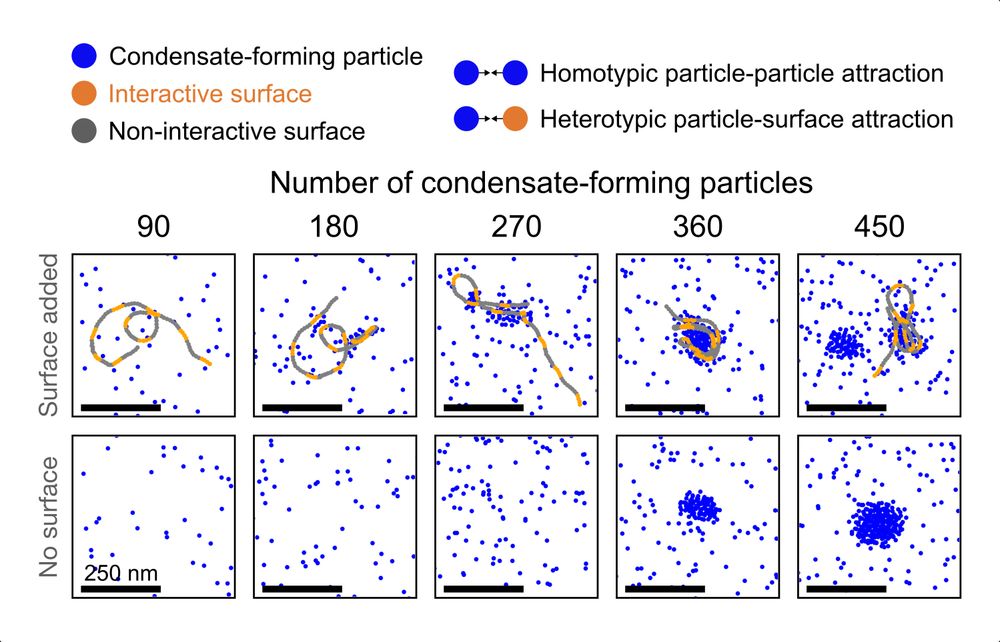
We are hiring! "Researcher in DNA-Based Information Processing Systems" (PhD or postdoc level). Co-supervision by Prof. Ralf Reussner (KIT computer science) and myself (biological systems). Combination of experiments and computational work. Please Apply / Repost!
www.pse.kit.edu/english/karr...
Exited to see SPIDR finally coming out! Congrats to Marko and the team on opening the next multiplexing RNA frontier 💥
23.07.2025 22:47 — 👍 3 🔁 0 💬 0 📌 0
Thrilled to co-advertise this postdoc or PhD position with @erhardtlab.bsky.social!
Join us, if RNP assembly on centromeres, RNA modifications, & bioinformatics are your thing.
More info also on modiclab.org.
Grateful for the fantastic collaboration with @mardakheh.bsky.social's lab and huge kudos to Anja, @jonanovljan.bsky.social, Valter, Martin, and everyone involved.
02.06.2025 10:49 — 👍 1 🔁 0 💬 0 📌 0
New method drop:
HCR-Proxy, a modular proximity labelling approach to profile local proteome composition around RNA at nanoscale, subcompartmental resolution. Thereby we resolved nested nucleolar layers and uncovered the grammar of spatial protein partitioning.
www.biorxiv.org/content/10.1...
Harnessing the physical properties of peptides to predict their functionality and interactions. 💥
24.05.2025 09:47 — 👍 3 🔁 0 💬 1 📌 0
Time for a short thread! We developed HiddenFoot, a biophysics-inspired approach to decode single-molecule footprinting data and infer TF, nucleosome, and RNA Pol II binding profiles on individual DNA molecules. One molecule at a time! www.biorxiv.org/content/10.1...
1/6
Thank you, Lennart! I'm very excited to join the newly founded Carl Zeiss Center for Synthetic Genomics and KIT with your lab nextdoor! Exciting times ahead!
18.04.2025 19:25 — 👍 6 🔁 2 💬 1 📌 0
I'm excited to share our latest publication on co-transcriptional RNA folding in a eukaryote! Great work by Leo Schärfen with collaboration from Isaac Vock and Matt Simon! @leoschaerfen.bsky.social @mattdsimon.bsky.social
Here is the link that works! authors.elsevier.com/a/1kpxo_Oylr...

The eternal question: should you focus on one problem for a very long time or is it more productive to bounce and work on many problems? David Baker says that both modes have been important in his career trajectory.
podcasts.apple.com/us/podcast/n... @nightsciencepod.bsky.social

📣 Our lab is looking for a PhD student to join us in exploring RNA biology! Details in the announcement. Feel free to reach out with any questions—and please RT!
25.03.2025 20:00 — 👍 8 🔁 12 💬 0 📌 1
Beautiful read from @gabrijelad.bsky.social, outlining what we today know and also have no clue about the exciting world of RNA localisation
genomebiology.biomedcentral.com/articles/10....

Save the date for the next edition of this wonderful meeting. We look forward to seeing you in Portugal!!
28.02.2025 15:53 — 👍 57 🔁 24 💬 2 📌 1
"Imagine a DAPI-like stain, but for the extracellular matrix." That's basically how this work was pitched to me by Kayvon and Antonio a year or so ago. Now the final product really delivers. Read about their versatile label for ECM in living tissues here: www.nature.com/articles/s41...
06.02.2025 16:02 — 👍 354 🔁 104 💬 9 📌 7Shoutout to the incredible team behind the integration of explainable #deeplearning (@jonanovljan.bsky.social), network based analysis (Ira Iosub) and tailored transcriptomics (Tajda Klobučar)
06.02.2025 09:44 — 👍 1 🔁 0 💬 0 📌 0
I am thrilled to share the first manuscript from my group on identifying sequence and structural features promoting the condensation of RNAs that are organising highly interconnected RNA networks, thereby expanding our understanding of higher-order transcriptome assemblage.
shorturl.at/pmLIj #RNASky