Big congrats to Clemens Steinek, @ipachmayr.bsky.social, and Sebastian Strauss who led the project as well as other co-authors that contributed to this work!! 6/6
07.02.2026 08:52 — 👍 1 🔁 0 💬 0 📌 0
Importantly, this workflow is broadly applicable and compatible with virtually any high off-rate binder. Many “bad” binders currently sitting in lab freezers may now become powerful imaging tools. 5/6
07.02.2026 08:52 — 👍 1 🔁 0 💬 1 📌 0

Using DyBE, we resolved the organization of receptor tyrosine kinases at single-protein resolution and detected ligand-dependent homodimerization of HER2, as well as EGF-induced formation of EGFR homodimers and EGFR–HER2 heterodimers. 4/6
07.02.2026 08:52 — 👍 1 🔁 0 💬 1 📌 0

DyBE increases protein labeling up to 15-fold for high off-rate nanobodies, enabling visualization of most molecules of a given target protein within the cell. 3/6
07.02.2026 08:52 — 👍 1 🔁 0 💬 1 📌 0

In classical DNA-PAINT, small binders such as nanobodies can localize proteins with nanometer precision, but rapid unbinding often limits efficient protein labeling. DyBE adapts DNA-PAINT to harness small, high off-rate binders for nanometer-precise sampling. 2/6
07.02.2026 08:52 — 👍 2 🔁 0 💬 1 📌 0

Here we introduce Dynamic Binder Exchange (DyBE), a new strategy that uses the high off-rate kinetics of small binders to map proteins with nanometer-scale precision! 1/6
07.02.2026 08:52 — 👍 1 🔁 0 💬 1 📌 0
How do you scale super-resolution microscopy to dozens of proteins without linearly scaling imaging time?
We introduce Combi-PAINT: a combinatorial DNA-PAINT strategy that breaks the 1-target-per-round bottleneck of Exchange-PAINT: www.biorxiv.org/content/10.6...
1/5
26.01.2026 09:34 — 👍 10 🔁 3 💬 1 📌 1

Ralf Jungmann, of the @mpibiochem.bsky.social, is the first speaker to step to the BiOS Hot Topics stage at #PhotonicsWest!💡
He is delivering his presentation titled: “From DNA nanotechnology to biomedical insight: towards single-molecule spatial omics”
@jungmannlab.bsky.social
18.01.2026 03:57 — 👍 4 🔁 1 💬 1 📌 0

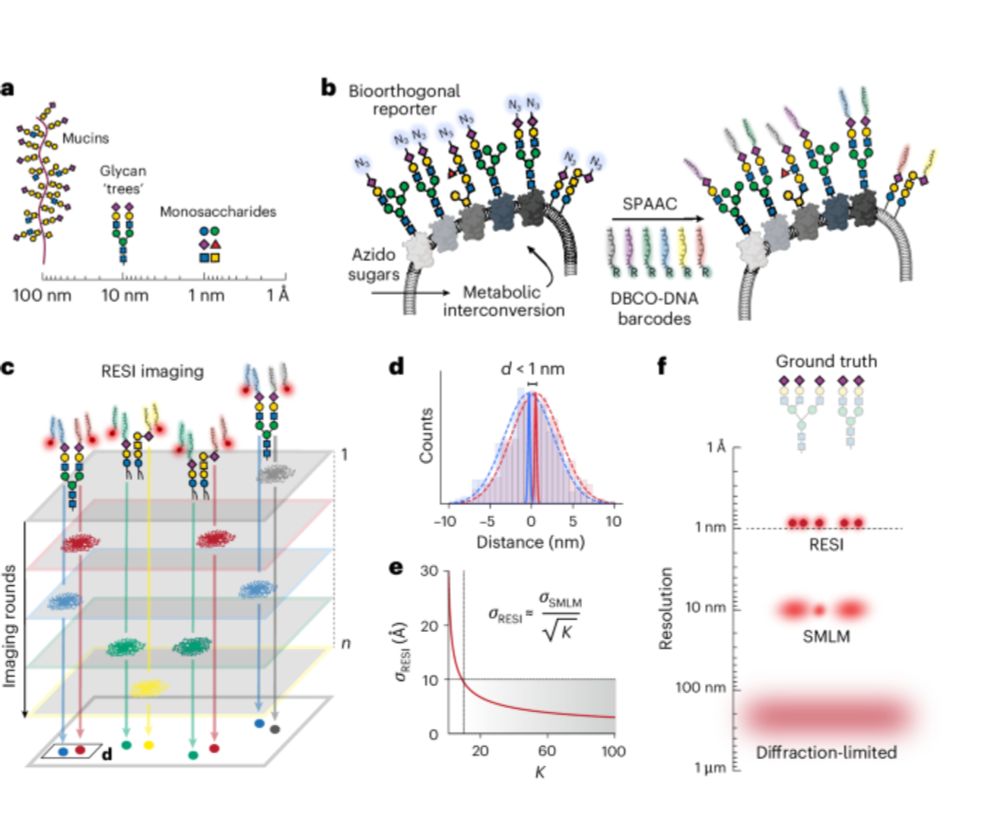
The image on the cover shows two sugars from the same cell-surface glycan separated by 9 Å, visualized with RESI (resolution enhancement by sequential imaging) enabled by metabolic labelling with DNA barcodes.
IMAGE: Luciano A. Masullo, Max Planck Institute of Biochemistry, Germany.
COVER DESIGN: Vanitha Selvarajan
Original paper: Masullo, L.A., et al. Ångström-resolution imaging of cell-surface glycans. Nat. Nanotechnol. 20, 1457–1463 (2025). https://doi.org/10.1038/s41565-025-01966-5
Abstract: Glycobiology is rooted in the study of monosaccharides, ångström-sized molecules that are the building blocks of glycosylation. Glycosylated biomolecules form the glycocalyx, a dense coat encasing every human cell with central relevance—among others—in immunology, oncology and virology. To understand glycosylation function, visualizing its molecular structure is fundamental. However, the ability to visualize the molecular architecture of the glycocalyx has remained challenging. Techniques such as mass spectrometry, electron microscopy and fluorescence microscopy lack the necessary cellular context, specificity and resolution. Here we combine resolution enhancement by sequential imaging with metabolic labelling, enabling the visualization of individual sugars within glycans on the cell surface, thus obtaining images of the glycocalyx with a spatial resolution down to 9 Å in an optical microscope.
Now online: October 2025 Issue.
- Focus Issue on #biosensing,
- DNA moiré superlattices,
- Sugars at Ångström-resolution,
- Solid-state #nanopores,
- Non-aqueous Li #batteries, -
- Neuromorphic vision,
- Peptide #hydrogels,
- Deep learning for #LNPs and more...
www.nature.com/nnano/volume...
17.10.2025 18:45 — 👍 8 🔁 5 💬 0 📌 0
Thanks to all who made this possible! @eduardunterauer.bsky.social @evaschentarra.bsky.social @ipachmayr.bsky.social @taishatashrin.bsky.social Jisoo Kwon Sebastian Strauss, @jekristina.bsky.social @rafalkowalew.bsky.social @opazo.bsky.social @forna.bsky.social @lumasullo.bsky.social (6/6)
02.10.2025 11:37 — 👍 5 🔁 1 💬 0 📌 0

Within this neuronal atlas we can reveal the three synapse classes, excitatory, inhibitory and the recently discovered mixed synapse. Organelle imaging of Peroxisomes (Pmp70) and the Golgi Apparatus (Golga5) reveals rare contact sides and even fused particles. (5/6)
02.10.2025 11:37 — 👍 7 🔁 1 💬 1 📌 0

To show the power of the technique, we acquired a 13-plex 200 x 200 µm2 neuronal atlas in 3D. With this atlas we map the interaction architecture of three neurons, resolving organelles, cytoskeleton, vesicles and synapses at single-protein resolution. (4/6)
02.10.2025 11:37 — 👍 5 🔁 1 💬 1 📌 0

We demonstrate speed-optimized left-handed DNA-PAINT by characterizing the sequence binding kinetics and resolving three main microscopy benchmarking targets, mitochondria, microtubules and nuclear pore complexes with <5 nm localization precision. (3/6)
02.10.2025 11:37 — 👍 7 🔁 1 💬 1 📌 0

The mirrored design of left-handed oligonucleotides allows the extension of the common 6 speed-sequences R1-R6 with their analogs L1-L6, enabling 12 target multiplexing with a standard secondary label-free DNA-PAINT workflow. (2/6)
02.10.2025 11:37 — 👍 6 🔁 1 💬 1 📌 0
Highly efficient 12-color multiplexing with speed-optimized DNA-PAINT. We are excited to share our latest paper in @natcomms.nature.com, using left-handed DNA to extend speed-optimized DNA-PAINT to 12 targets in a simple and straightforward way! 🧬👈🚀https://www.nature.com/articles/s41467-025-64228-x
02.10.2025 11:37 — 👍 28 🔁 9 💬 2 📌 2


Next on stage is Eduard Unterauer @eduardunterauer.bsky.social from @jungmannlab.bsky.social reporting spatial proteomics with DNA PAINT #SMLMS2025
27.08.2025 12:17 — 👍 19 🔁 5 💬 0 📌 0
We're excited that the study is now out in Nature Nanotechnology @natnano.nature.com www.nature.com/articles/s41...
30.07.2025 14:10 — 👍 6 🔁 0 💬 0 📌 0
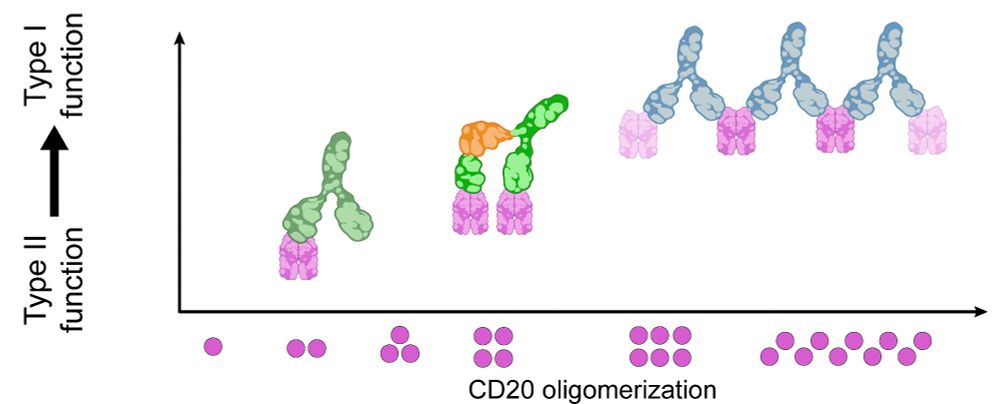
The shift from Type II to Type I function reveals a structure–function continuum for anti-CD20 antibodies, showing that receptor arrangements dictate mechanism of action. RESI provides a platform for structure-guided antibody development, applicable far beyond CD20. (5/6)
28.07.2025 08:36 — 👍 3 🔁 0 💬 1 📌 0
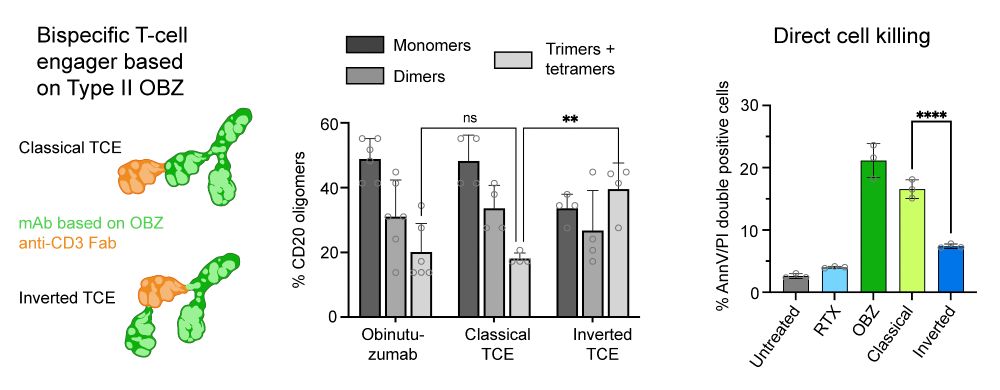
We showed a direct link between CD20 oligomerization and function by investigating OBZ-based T-cell engagers (TCEs). An increased IgG flexibility in the 2+1 TCE format lead to increased CD20 tetramerization, without higher-order clustering, resulting in a reduction of direct cytotoxicity. (4/6)
28.07.2025 08:36 — 👍 4 🔁 0 💬 1 📌 0
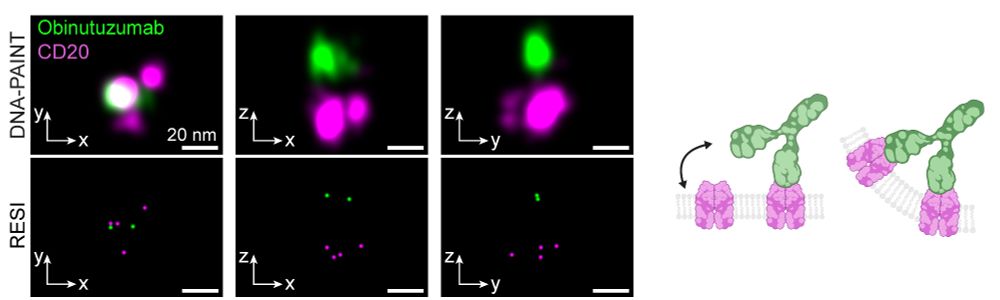
In contrast, Type II antibodies like Obinutuzumab and H299 induced limited oligomerization to dimers, trimers and tetramers, consistent with their role in promoting direct tumor cell death rather than complement activation. (3/6)
28.07.2025 08:36 — 👍 3 🔁 0 💬 1 📌 0
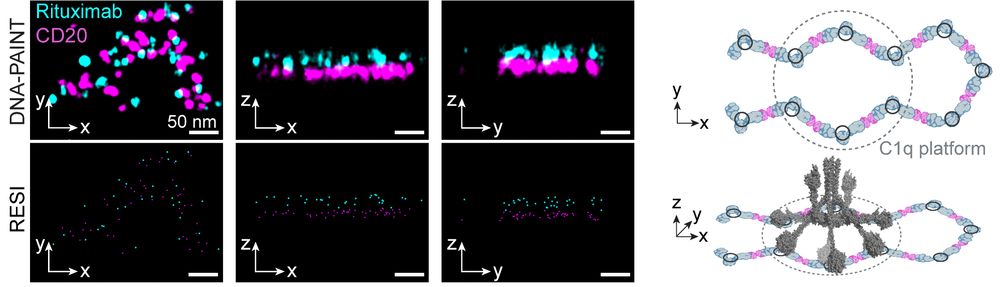
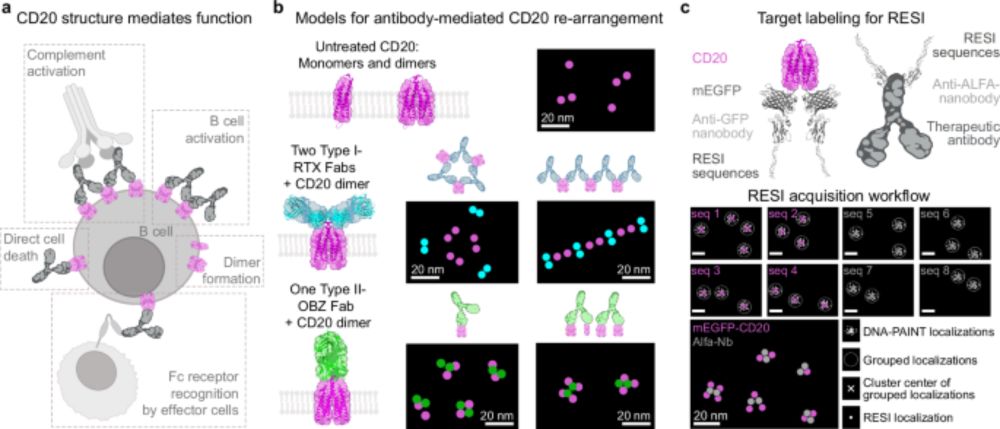
By imaging intact cells, we could see these therapeutic antibodies in action: Type I antibodies like Rituximab and Ofatumumab formed extended chains of CD20 hexamers or larger, creating platforms compatible with complement protein binding, mediating cancer cell killing. (2/6)
28.07.2025 08:36 — 👍 3 🔁 0 💬 1 📌 0
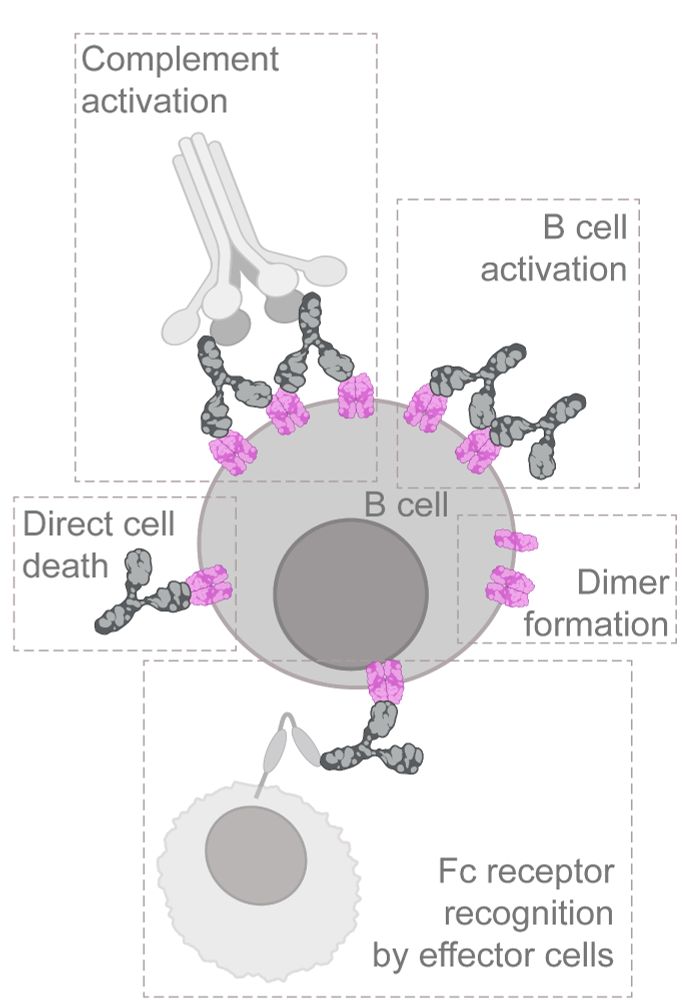
Ever wondered what happens when therapeutic antibodies bind to cancer cells? In our latest study, we used multiplexed 3D-RESI to directly visualize how anti-CD20 antibodies interact with their receptors, revealing their precise arrangement at single-protein resolution. (1/6)
28.07.2025 08:36 — 👍 24 🔁 7 💬 1 📌 0

Congratulations to Ralf on your election as a new EMBO member:
❕Original press release from @embo.org : www.embo.org/press-releas...
03.07.2025 08:57 — 👍 14 🔁 2 💬 0 📌 0

Great science, great company and stunning views at our Lab retreat on Schloss Ringberg 🏰🧬🔬. Big thanks to our guests Sabrina Simoncelli, Sebastian Kobold, Thomas Schlichthärle, @massivephotonics.bsky.social & students from the @lfmilles.bsky.social and @mlsb-borgwardt.bsky.social Labs for joining!
14.06.2025 18:31 — 👍 14 🔁 2 💬 0 📌 0
@larissaheinze.bsky.social
07.05.2025 15:21 — 👍 1 🔁 0 💬 0 📌 0
@moniquehonsa.bsky.social , @philippsteen.bsky.social , Larissa Heinze, Shuhan Xu, Heinrich Grabmayr, Isabelle Pachmayr, Susanne C. M. Reinhardt, Ana Perovic, Jisoo Kwon, Ethan P. Oxley, Ross A. Dickins, Maartje M. C. Bastings, Ian A. Parish
07.05.2025 14:42 — 👍 2 🔁 0 💬 1 📌 0
Max Planck Institute investigating the structure and function of proteins - from individual molecules to whole organisms.
Center of excellence for the training of doctoral researchers in the areas of biochemistry 👩🔬, structural biology 🧬 , biophysics ⚛️, cell biology 🧫, systems biology 🔬, and computational biology 💻.
https://imprs-ml.mpg.de/
Interdisciplinary research center at LMU Munich
Physicist - Doctoral Candidate @jungmannlab.bsky.social
Exploring the cellular world at the nanometer scale with DNA-PAINT.
@mpibiochem.bsky.social
@lmumuenchen.bsky.social
PhD student, microscopy, analysis, DNA-PAINT, SMLM.
@jungmannlab.bsky.social, Munich, MPI of Biochemistry.
Tinnefeld lab @lmumuenchen.bsky.social 🧬
In our lab, hardcore biochemistry meets physics. And coffee. We are investigating DNA replication through single molecule loupe @ Technical University of Munich and MPI of Biochemistry
Starting Lab at GeneCenter (LMU Munich) and MPI of Biochemistry, Emmy Noether Programme, former postdoc @UWproteindesign
We systematically elucidate the mechanisms underlying cell division and nuclear organisation and develop advanced imaging technologies
@EMBL.org
Physicist | PhD Student @JungmannLab @MPI_Biochem @LMU_Muenchen
Super-resolution microscopy 🔬🧬 Spatial-omics technology 👨🎨🌈 Neuroscience Applications 🧠
Physics PhD student in the JungmannLab (MPI_Biochem / LMU_Muenchen / IMPRS_ML).
Pushing the limits of microscopy!
Biotechnologist - PhD student @jungmannlab.bsky.social
Enjoying the little things in life using #DNAPAINT 🔬🧬
@mpibiochem.bsky.social
@lmumuenchen.bsky.social
@imprs-ml.bsky.social
Spatial Omics | Super-resolution imaging | DNA Nanotechnology | #DNAPAINT #superresolution #spatialomics #microscopy Professor
@schuederlab @EPFL
Physicist | single molecules, bioscience, biotech, nanotech, optics, fluorescence | Project Leader at @jungmannlab.bsky.social Max Planck Institute of Biochemistry @mpibiochem.bsky.social
THE DNA-PAINT COMPANY. We offer comprehensive DNA-PAINT labeling kits & DNA-PAINT solutions such as services and collaborations. #SMLM #superresolution #DNAPAINT #microscopy