
Clustering hundreds of millions of molecules in a single workstation is now a possibility!
Check the new code: github.com/mqcomplab/bb...
@evasmorodina.bsky.social
Computational structural biologist

Clustering hundreds of millions of molecules in a single workstation is now a possibility!
Check the new code: github.com/mqcomplab/bb...
Very nice!
05.09.2025 10:07 — 👍 2 🔁 1 💬 0 📌 0
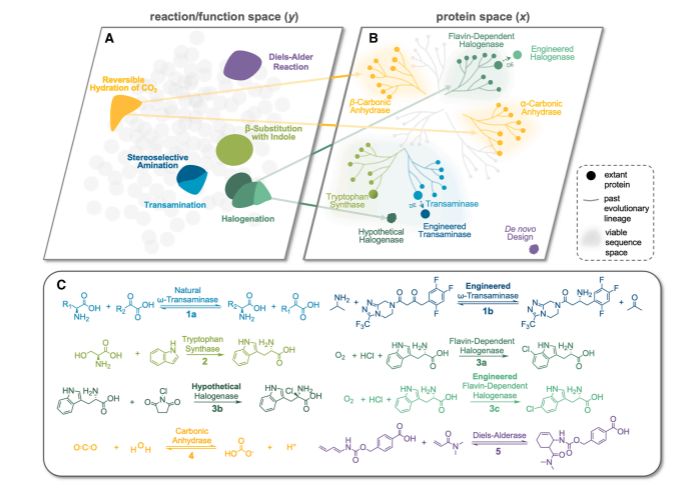
A compelling review of how ML/AI could help in the quest to find an enzyme for every reaction.
@jsunn-y.bsky.social @francescazfl.bsky.social Yueming Long @francesarnold.bsky.social
www.cell.com/cell-systems...

Preprint: De-novo design of proteins that inhibit bacterial defenses
Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵
Glad to share the final version of our story about the UBR4 complex, an E4 ligase protein quality control hub @science.org. Now with more cryo-EM structures and a deeper dive into substrate recognition, especially escaped mitochondrial proteins @clausenlab.bsky.social www.science.org/doi/10.1126/...
28.08.2025 18:54 — 👍 80 🔁 30 💬 7 📌 1
#CCPBioSim will be running their annual training week of hands-on workshops in basic biomolecular simulation techniques from the 13th-17th October, in Sheffield.
For more details, visit www.ccpbiosim.ac.uk/training2025
The registration link there will open shortly! #compchem
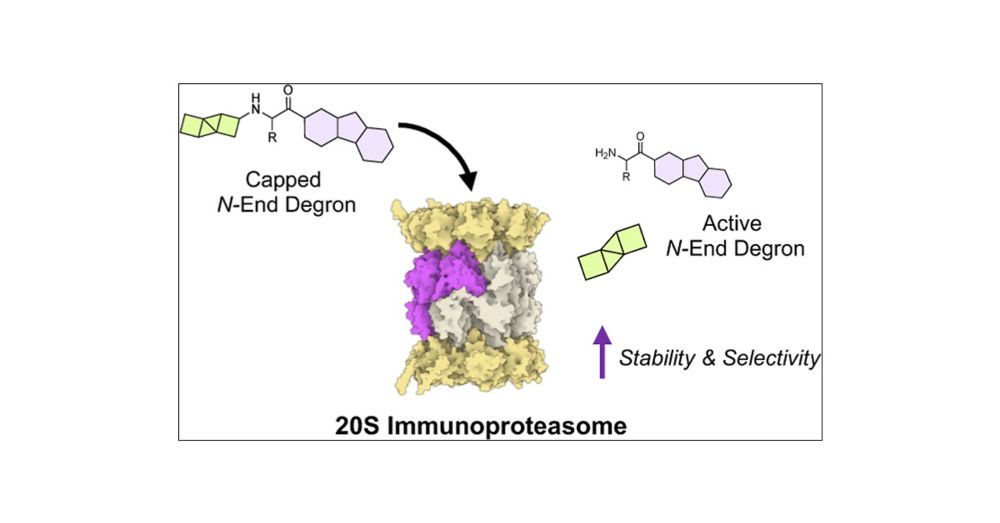
New degrader paper using the immunoproteasome. Congrats Cody Loy and Tim Harris!
pubs.acs.org/doi/full/10....
encapsulate your target proteins for cryoEM structural determination within highly hydrophilic, structurally homogeneous, and stable protein shells 🤩
www.biorxiv.org/content/10.1...

We're excited to present our latest article in Nature Machine Intelligence - Boosting the predictive power of protein representations with a corpus of text annotations.
Link: www.nature.com/articles/s42...
[1/4]

CACHE 7 is launched with support from the @gatesfoundation.bsky.social and unpublished data from Damian Young at @bcmhouston.bsky.social, Tim Willson @thesgc.bsky.social and Neelagandan Kamaria InSTEM. Design selective PGK2 inhibitors. We'll test them experimentally.
bit.ly/4lnVYOs
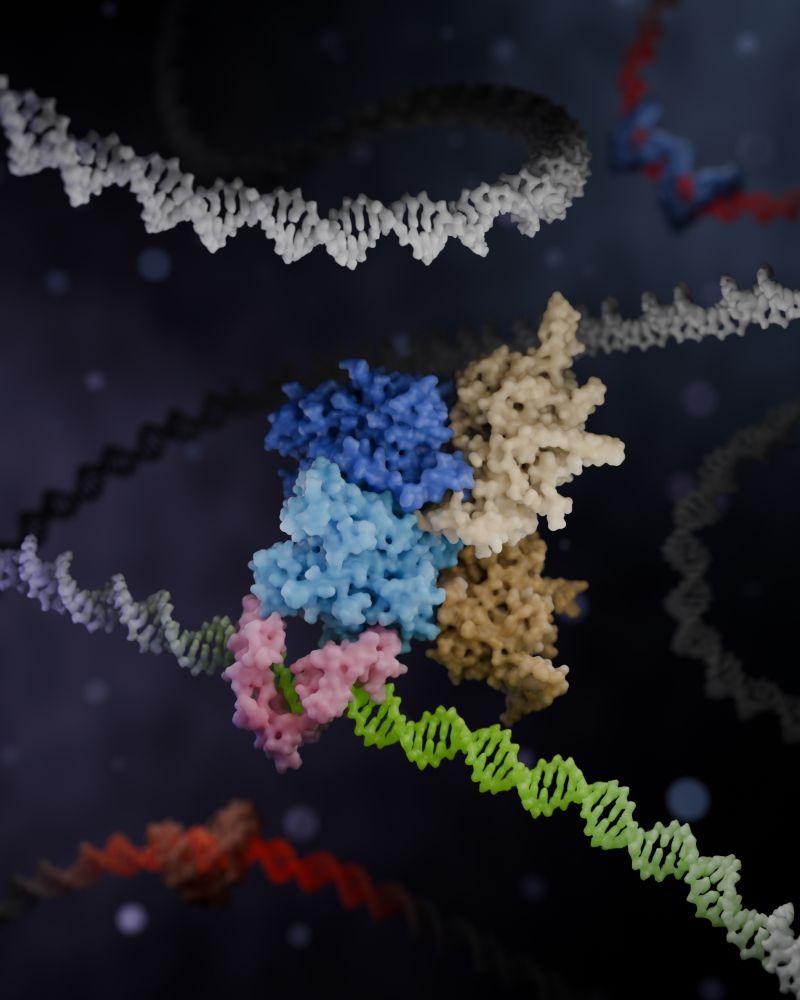
3D render of a Zincore complex locking ZFP91 onto DNA, promoting gene activation. Made in Blender with Molecular Nodes.
📢 Ready for this? A new way to think about Gene Regulation!
🚨A new coregulator complex, Zincore, acts as a "MOLECULAR GRIP", stabilizing TFs at DNA binding sites across the genome!🧬
💪Proud to be part of this work led by @daaninthelab.bsky.social
Read more: science.org/doi/10.1126/science.adv2861
🚀 My first Cryo-EM structures are OUT on the PDB!
🎉Super excited about this milestone as PhD, it’s been a journey, and I’m grateful to finally have these as my first Cryo-EM structures! 🙌
🔁Thank you for sharing!❤️
Read @daaninthelab.bsky.social et. al.: www.science.org/doi/10.1126/...
⬇️ 2nd video
🚨It’s out @embojournal.org 🚨
Cardiolipin inhibits Caspase-4/11!
Huge thanks to my brilliant supervisors Kate & Pablo, to @s-burgener.bsky.social & Mercedes and all the @inflammasomelab.bsky.social, it was a joy working and laughing with you, and to @brozlab.bsky.social & Si Ming lab.

We have a new collection of protein structure generative models which we call Protpardelle-1c. It builds on the original Protpardelle and is tailored for conditional generation: motif scaffolding and binder generation.
19.08.2025 17:16 — 👍 22 🔁 5 💬 1 📌 1
Switching next to protein protein interactions, we used arguably my favorite protein A34F GB1 to demonstrate that these glycopolymers stabilize protein protein interactions in a similar fashion to protein folding, via chemical interactions.
13.08.2025 17:25 — 👍 7 🔁 3 💬 1 📌 0
Interested in solvation free energies of small molecules and proteins?
The latest @livecomsjournal.bsky.social tutorial by Egger-Hoerschinger et al provides a guide to quantifying hydration thermodynamics using Grid Inhomogeneous Solvation Theory (GIST):
doi.org/10.33011/liv...
#compchem

Pushing down to the structures of ever-smaller proteins (and ever-smaller crystals!)
13.08.2025 21:43 — 👍 40 🔁 10 💬 3 📌 1Happy to share the latest from the lab, led by Daniel Alvarez, in collaboration with @lizconibear.bsky.social. In this AA-MD tour-de-force, we delve deep into the mechanism and energetics of lipid uptake by bridge-like lipid transfer proteins, and we learn a few interesting things along the way...
07.08.2025 08:45 — 👍 42 🔁 14 💬 1 📌 0Thrilled to share my first corresponding author paper as a project leader in @hillerlab.bsky.social Amazing work from first author @annaleder.bsky.social We discovered multi-chaperone condensates in the ER that revolutionise the current vision of protein folding!
www.nature.com/articles/s41...
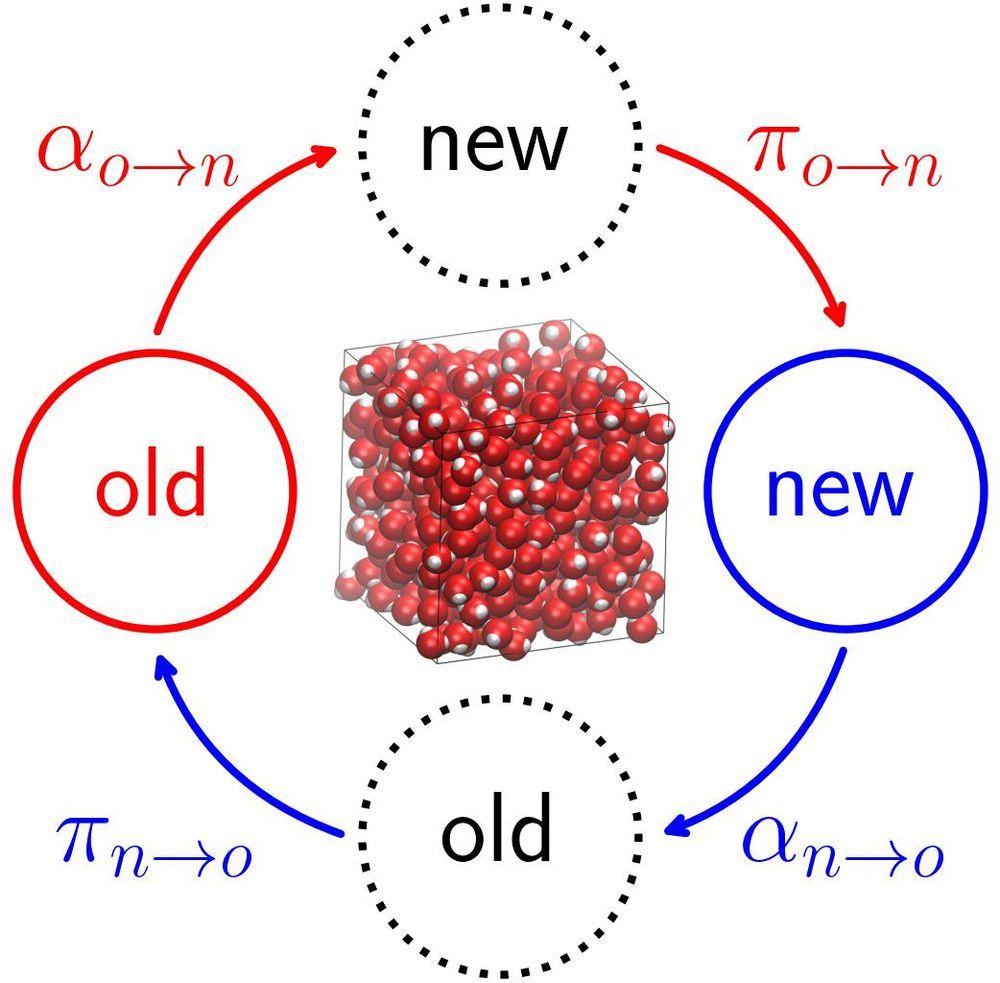
Table of contents figure
Our latest Best Practices article "Developing Monte Carlo Methodologies in Molecular Simulations" is out now and describes how to derive acceptance probabilities for a variety of Monte Carlo moves:
doi.org/10.33011/liv...
#compchem
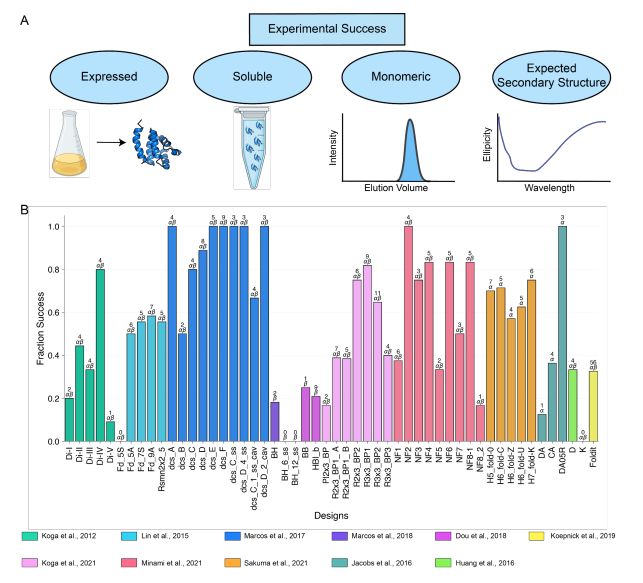



A benchmark dataset of 614 experimentally characterized de novo designed monomers from 11 different design studies shows that:
- deep learning structural metrics only weakly predict success
- The score distribution is different for different types of structures
@grocklin.bsky.social
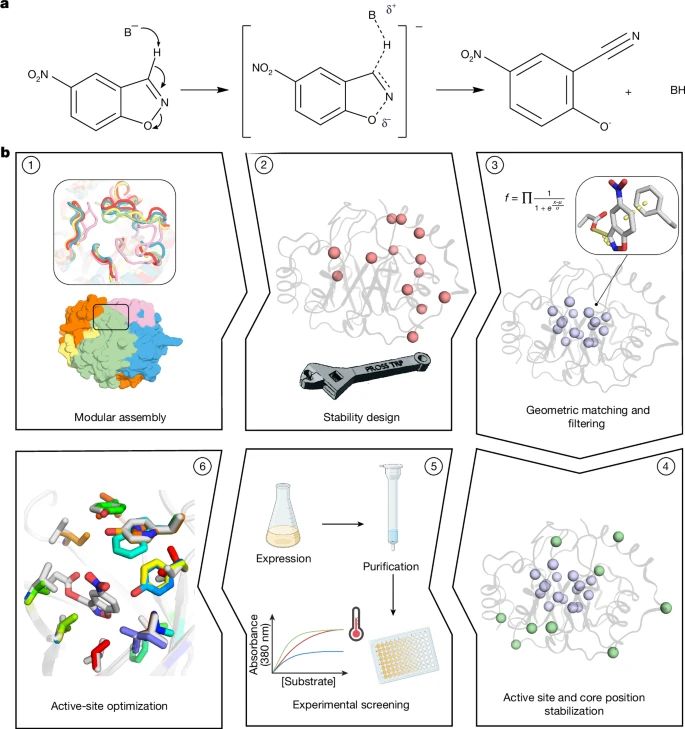
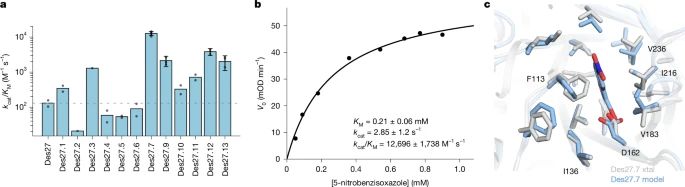
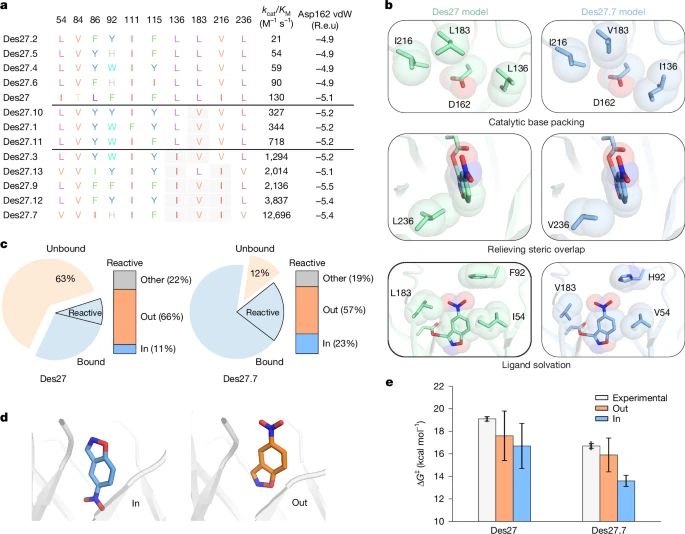
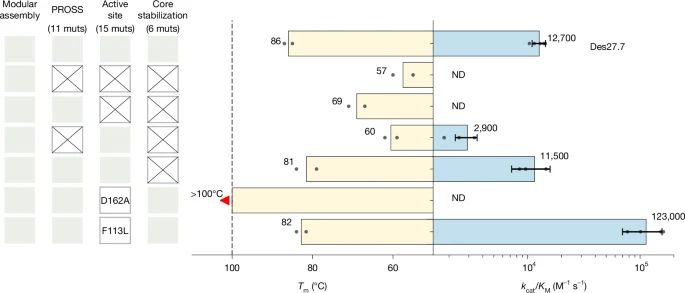
Physics-based design of efficient Kemp eliminases
@lynnkamerlin.bsky.social
www.nature.com/articles/s41...
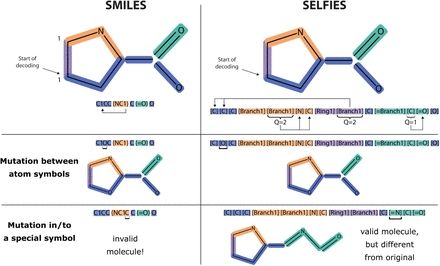
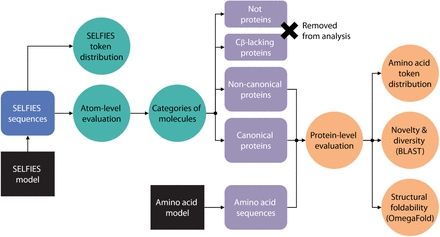

All all-atom diffusion model of protein sequences.
www.biorxiv.org/content/10.1...
Very excited about our latest all-atom generative model proteina, check out the project page (research.nvidia.com/labs/genair/...) and stay tuned for the code release soon!
19.07.2025 20:46 — 👍 24 🔁 9 💬 0 📌 1
Transferring Knowledge from MM to QM: A Graph Neural Network-Based Implicit Solvent Model for Small Organic Molecules | Journal of Chemical Theory and Computation pubs.acs.org/doi/10.1021/...
04.08.2025 13:20 — 👍 4 🔁 1 💬 0 📌 0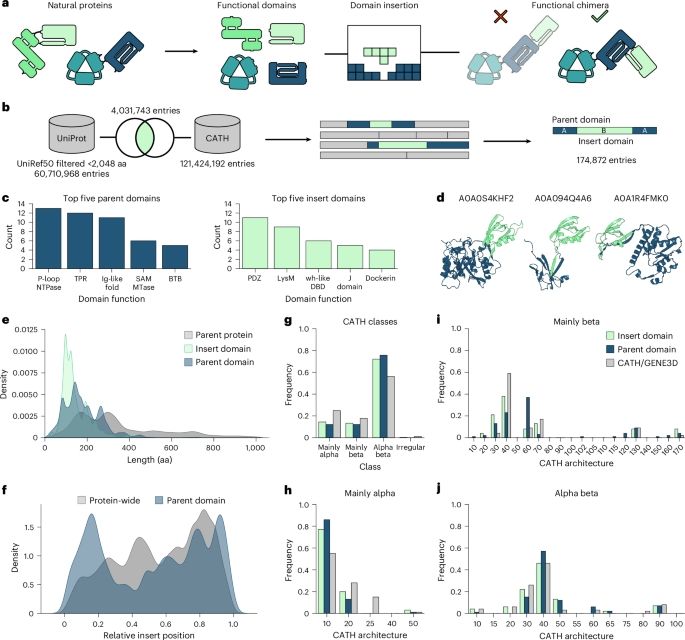
Our work on rationally engineering allosteric protein switches is now out in Nature Methods: www.nature.com/articles/s41... Thanks a lot to @grunewald.bsky.social and @noahholzleitner.bsky.social for the comprehensive news and views: www.nature.com/articles/s41...
04.08.2025 11:41 — 👍 25 🔁 8 💬 0 📌 0
Excited to share work with
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
One for the Apicomplexa peeps:
ever wonder how does the IMC grow so fast during progeny formation? Where do all the lipids come from? In our latest work on malaria RBC stages we implicate this monster protein (~6000 residues!) and ER contact sites.
Happy+proud to share and keen to hear thoughts!
If you’re interested in learning more about protein folding and misfolding, I’ve created a convenient reading list with a few essential papers:
scholar.google.com/citations?us...
scholar.google.com/citations?us...
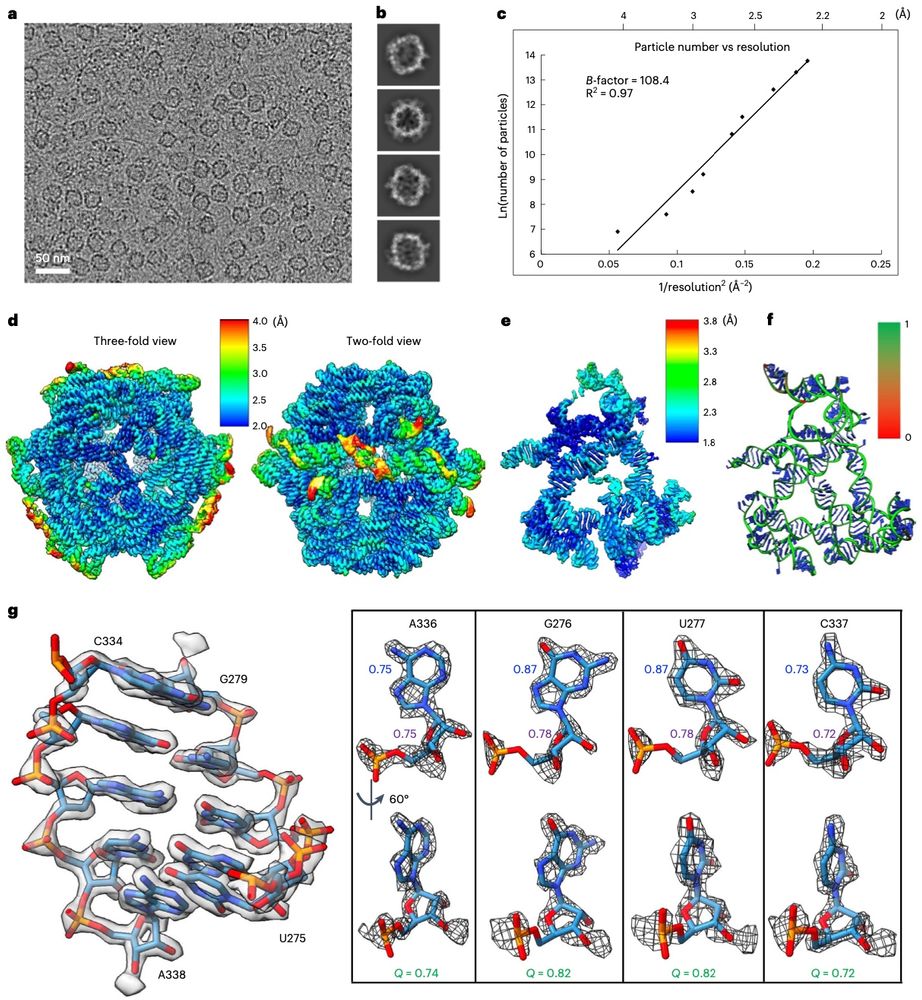
High resolution CryoEM maps of RNA assemblies
Not my area at all, but how cool are these cryoEM structures of purely RNA-based assemblies!
www.nature.com/articles/s41...