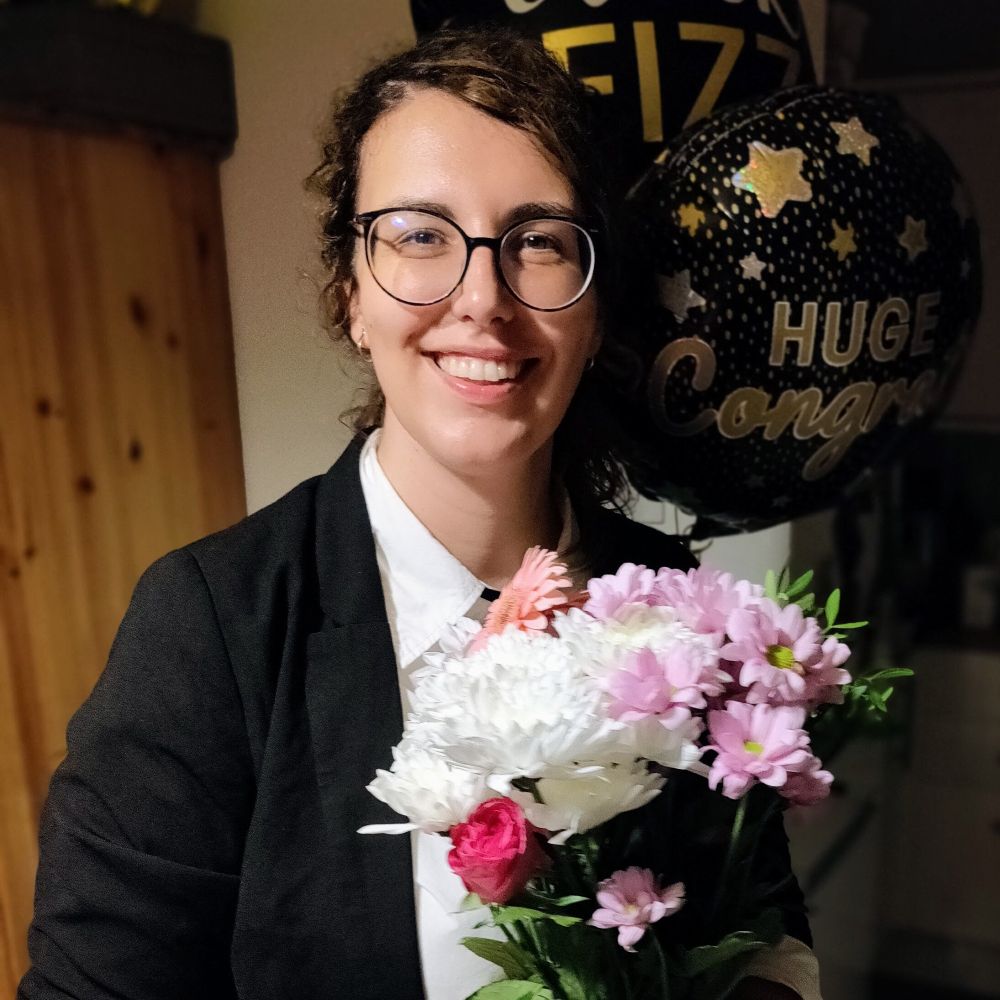Deadline extended to August 1st!
Fully funded DPhil (PhD) in Statistical Genetics at Oxford, available to international candidates!
21.07.2025 11:39 — 👍 1 🔁 3 💬 0 📌 0
Decoding cnidarian cell type gene regulation
Animal cell types are defined by differential access to genomic information, a process orchestrated by the combinatorial activity of transcription factors that bind to cis -regulatory elements (CREs) to control gene expression. However, the regulatory logic and specific gene networks that define cell identities remain poorly resolved across the animal tree of life. As early-branching metazoans, cnidarians can offer insights into the early evolution of cell type-specific genome regulation. Here, we profiled chromatin accessibility in 60,000 cells from whole adults and gastrula-stage embryos of the sea anemone Nematostella vectensis. We identified 112,728 CREs and quantified their activity across cell types, revealing pervasive combinatorial enhancer usage and distinct promoter architectures. To decode the underlying regulatory grammar, we trained sequence-based models predicting CRE accessibility and used these models to infer ontogenetic relationships among cell types. By integrating sequence motifs, transcription factor expression, and CRE accessibility, we systematically reconstructed the gene regulatory networks that define cnidarian cell types. Our results reveal the regulatory complexity underlying cell differentiation in a morphologically simple animal and highlight conserved principles in animal gene regulation. This work provides a foundation for comparative regulatory genomics to understand the evolutionary emergence of animal cell type diversity. ### Competing Interest Statement The authors have declared no competing interest. European Research Council, https://ror.org/0472cxd90, ERC-StG 851647 Ministerio de Ciencia e Innovación, https://ror.org/05r0vyz12, PID2021-124757NB-I00, FPI Severo Ochoa PhD fellowship European Union, https://ror.org/019w4f821, Marie Skłodowska-Curie INTREPiD co-fund agreement 75442, Marie Skłodowska-Curie grant agreement 101031767
I am very happy to have posted my first bioRxiv preprint. A long time in the making - and still adding a few final touches to it - but we're excited to finally have it out there in the wild:
www.biorxiv.org/content/10.1...
Read below for a few highlights...
06.07.2025 18:14 — 👍 58 🔁 24 💬 1 📌 2

Machine Learning in Computational Biology
Youtube channel for the Machine Learning in Computational Biology conference.
Just under a week until the #MLCB2025 paper/abstract deadline on June 1st! In-person registration is full but you can join the wait list forms.gle/gnj6AAV7oWj6... or watch online at youtube.com/@mlcbconf. Sept 10-11 at @nygenome.org. Full deets at mlcb.org! Please RP.
26.05.2025 16:16 — 👍 10 🔁 4 💬 0 📌 0

Come and join us! We’re hiring a new Group Leader in Generative Biology at the @sangerinstitute.bsky.social
Building AI models or the data to train them?
Core funding of >$130M a year for a faculty of ~30.
www.nature.com/naturecareer...
acrobat.adobe.com/id/urn:aaid:...
pls RT!
06.05.2025 14:59 — 👍 46 🔁 67 💬 1 📌 0

Graph at ICLR 2025 in Singapore from 24 - 28 April. Meet us there!

A portrait of Katherine Driscoll, Head of AI at Graph, standing in front of a blackboard with mathematical formulas.
Hey Singapore, @kdris.bsky.social will be at ICLR 2025 from 24 - 28 April! We’re looking forward to hearing about the latest discoveries in biology, AI, and deep learning. Let us know if you’ll be around! 🇸🇬
22.04.2025 09:13 — 👍 5 🔁 2 💬 0 📌 0
I get "page not found" -- could you repost the link?
19.03.2025 01:27 — 👍 0 🔁 0 💬 1 📌 0

Ai2 ScholarQA logo
Can AI really help with literature reviews? 🧐
Meet Ai2 ScholarQA, an experimental solution that allows you to ask questions that require multiple scientific papers to answer. It gives more in-depth and contextual answers with table comparisons and expandable sections 💡
Try it now: scholarqa.allen.ai
21.01.2025 19:30 — 👍 33 🔁 12 💬 1 📌 6
📢 Calling all researchers! The #LMRL workshop is now accepting paper submissions! 🧬✨
🎯 Showcase complete works / early-stage ideas 💡 via your chance to inspire & connect with a non-traditional AIxBio #ICLR2025 audience 🌍
👉 Help us spread the word of lmrl.org 🙌
06.01.2025 15:00 — 👍 13 🔁 6 💬 0 📌 0

ICLR 2025 Workshop Proposals
Welcome to the OpenReview homepage for ICLR 2025 Workshop Proposals
The list of accepted workshops for ICLR 2025 is available at openreview.net/group?id=ICL...
@iclr-conf.bsky.social
We received 120 wonderful proposals, with 40 selected as workshops.
03.12.2024 16:29 — 👍 57 🔁 15 💬 1 📌 5
Three BioML starter packs now!
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
03.12.2024 03:27 — 👍 147 🔁 60 💬 16 📌 6
It’s already December 2nd, which means 2 Advent of Code 2024 puzzles are out! I’m sharing my solutions on my blog: kasia.codes/posts/aoc24/. If you’ve got a bit of time, highly recommend giving it a shot—it's quite fun! 🎄💻 adventofcode.com #AdventOfCode #aoc2024
02.12.2024 15:58 — 👍 2 🔁 0 💬 0 📌 0
Thank you for posting this. 😊 I saw this on the other app some time ago but didn't manage to bookmark it, and Google was no help.
24.11.2024 19:29 — 👍 2 🔁 0 💬 0 📌 0
Search Jobs | Microsoft Careers
In the spirit of trying to be more active on here... the deadline for our BioML internship is tomorrow 11/22.
If you are currently a PhD student please consider applying --- all we need is a CV and research statement!
jobs.careers.microsoft.com/global/en/jo...
21.11.2024 17:57 — 👍 6 🔁 3 💬 1 📌 1
Two BioML starter packs now:
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
DM if you want to be included (or nominate people who should be!)
18.11.2024 17:09 — 👍 119 🔁 58 💬 10 📌 11
The @bsky.app is feeling more and more like the lost #sci-Twitter and I am all for it! 🤞 Missed the science space and Twitter or X hasn't been that ever since the sink incident. #exodus #science #twitter
18.11.2024 01:44 — 👍 1 🔁 0 💬 0 📌 0
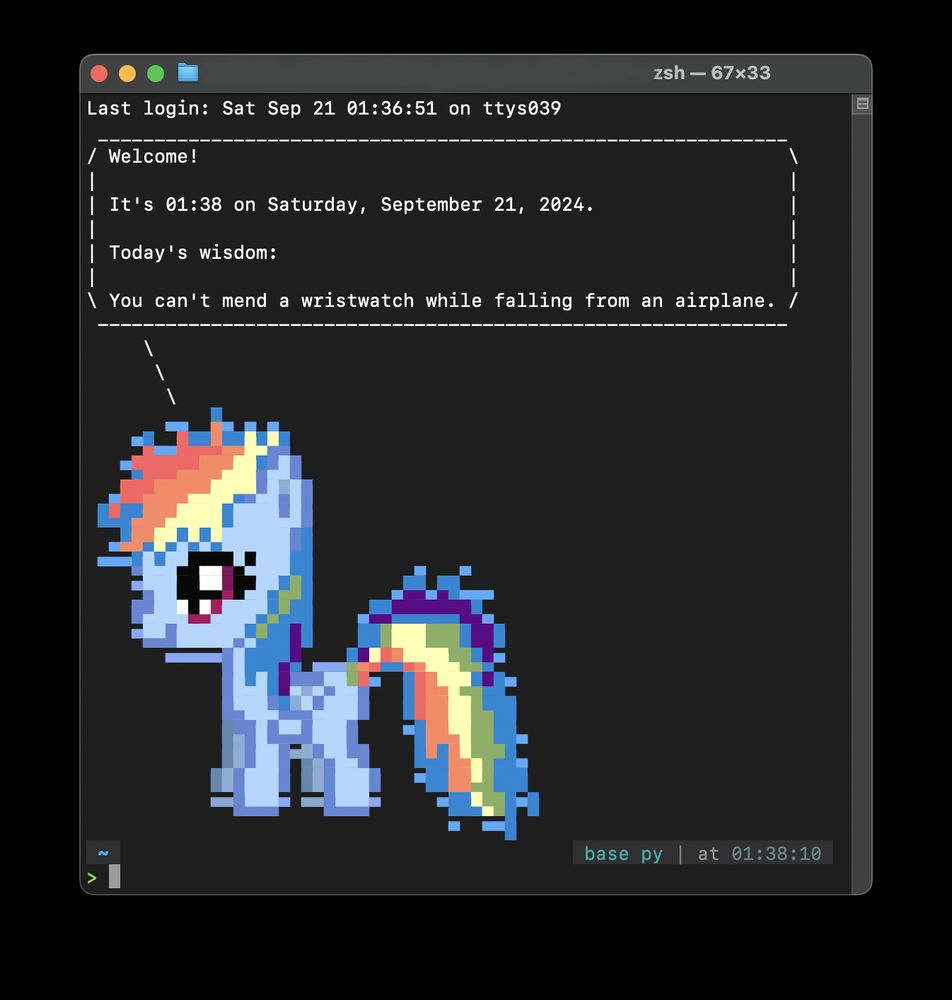
Screenshot of a terminal session showing a greeting message with the current date, time, and a piece of wisdom: 'You can't mend a wristwatch while falling from an airplane.' Below the text, there's a colorful ASCII art depiction of a pony with a rainbow mane and tail.
✨ Brighten up your terminal! ✨
Who wouldn’t want to be greeted by a pony 🐎 or a cow 🐄 during in their terminal?! 🙈
I wrote a post on adding some fun and color to your console: kasia.codes/posts/pony_i...
#bash #zsh #ponysay #cowsay #TerminalFun
17.11.2024 19:00 — 👍 1 🔁 0 💬 0 📌 0
who knew it would be easier to get everyone to switch social media platforms than getting them to use hg38
14.11.2024 18:45 — 👍 254 🔁 62 💬 15 📌 13
Here's hoping to having enough time to solve all 25x2 puzzles! 🤞
16.11.2024 13:31 — 👍 1 🔁 0 💬 0 📌 0

A festive holiday scene with a laptop displaying glowing code, surrounded by Christmas decorations like fairy lights, ornaments, a candy cane, and a small tree. Snowfall is visible through a window, creating a cozy atmosphere with subtle coding elements woven into the decor.
December's coming, and so is my favorite recent tradition: #AdventOfCode! 🎄👩💻 Excited to solve coding puzzles in the spirit of helping elves and saving holidays. Highly recommend! adventofcode.com
16.11.2024 13:30 — 👍 3 🔁 0 💬 1 📌 1
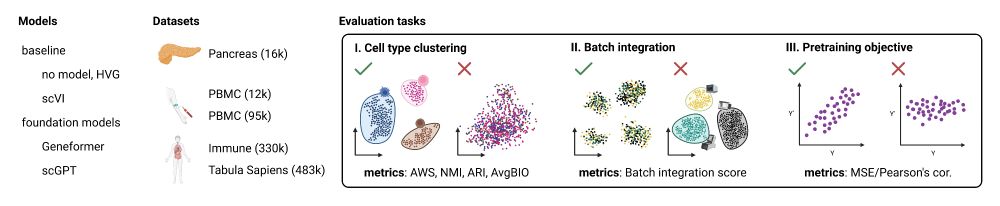
This is a summary figure of the evaluation setup where two foundation models, Geneformer and scGPT, together with baselines such as no model (HVG) and scVI were evaluated on 5 datasets in 3 tasks for cell type clustering, batch integration, and pretraining objective. The figure illustrates the expected good and bad cases for each task. In cell type clustering a good outcome is when cell types separate in the embedding space, while bad is when they are intermixed. In batch integration, expected is the separation across cell types, not batches. In the pretraining objective, a positive correlation is expected between input and predicted values.
How robust are proposed foundation models in single-cell biology?
Here, we assessed two models: Geneformer and scGPT. We focus on evaluating their zero-shot capabilities.
📄 biorxiv.org/content/10.1...
💻 github.com/microsoft/ze...
💪 with Alex Lu, Ava Amini, and Lorin Crawford
18.10.2023 15:43 — 👍 1 🔁 0 💬 0 📌 0
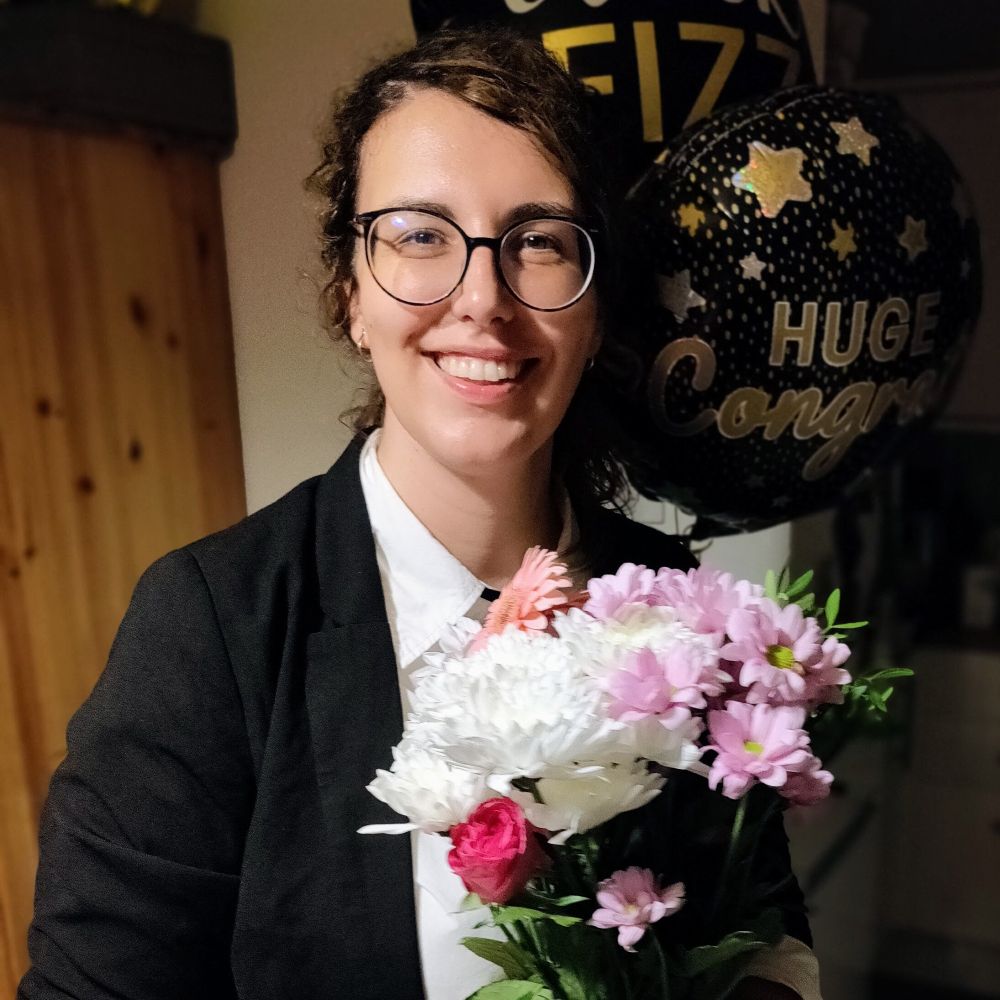
photo of me holding flowers with congratulations balloons behind.
Yay, I passed my viva today! 🎓💪
I had to take a photo home, as thanks to Oxford's weather, I only dried up after the pub. :) Had great fun celebrating and will take my well-deserved time off from my thesis (before submitting the corrections, of course). #phdone #phdchat
12.09.2023 22:08 — 👍 4 🔁 0 💬 0 📌 0
computational biologist. all things genomic. working on cfDNA, cancer genomics, early detection, prenatal testing, epigenetics, transposable elements. building reproducible computational pipelines. seeking academic partners for collaboration.
Scientist and Group Leader of the Simons Machine Learning Center
@SEMC_NYSBC. Co-founder and CEO of http://OpenProtein.AI. Opinions are my own.
Research Scientist AI/ML @cziscience.bsky.social , PhD @www.helmholtz-munich.de , OSS @scverse.bsky.social
giovannipalla.com
Computational biologist interested in deciphering the genomic regulatory code at vib.ai
Columnist and chief data reporter the Financial Times | Stories, stats & scatterplots | john.burn-murdoch@ft.com
— On 👨👶 leave until July —
📝 ft.com/jbm
Associate Professor of Bioinformatics at University of Tartu, Estonia. Project lead at eQTL Catalogue. https://kauralasoo.github.io/
Head of Data Science & Engineering at Talus Bio. I post about data science, BioML, AI drug discovery, Python, mass spec, proteomics, and silly things my kids do.
I also blog sometimes: https://willfondrie.com/post
Computer Science PhD Student at @humboldtuni.bsky.social and @mdc-berlin.bsky.social | Data Science | Machine learning | AI | Bioinformatics | Genomics | Single-Cell Biology
NCI K00 Fellow in Janes Lab at U of Virginia BME (USA) working on ncRNAs in breast cancer. PhD on ncRNAs in myogenesis in Dutta lab at UVA BMG. MSc from U of Warsaw (Poland). Mom of three and wife of Dr. Piotr Przanowski, scientist and HEMA fighter.
Breakthrough AI to solve the world's biggest problems.
› Join us: http://allenai.org/careers
› Get our newsletter: https://share.hsforms.com/1uJkWs5aDRHWhiky3aHooIg3ioxm
Understanding life. Advancing health.
Waitress turned Congresswoman for the Bronx and Queens. Grassroots elected, small-dollar supported. A better world is possible.
ocasiocortez.com
Co-founder & CSO @ArcadiaScience. Head of Open Science @AsteraInstitute. Immigrant. she/her
Still post frequently on X (@PracheeAC)
Created http://adventofcode.com, http://compute-cost.com, http://anoik.is, http://was.tl/projects/; Principal Architect at https://acvauctions.com
Spaceflight, Biology, Health, Clinical, Data, Informatics/AI/ML. As humans return🚀 to 🌘 1000s+ can engage via #OpenScience. I❤️Rugby. Personal acct/not work’s. GS: https://scholar.google.com/citations?user=Xy9lW6wAAAAJ&hl=en
Machine Learning & Systems Biology. ML Group Leader @arcinstitute. PhD @StanfordAILab
http://www.yusufroohani.com
R, data, 🐕, 🍸, 🌈. He/him.
Foundational tools for omics data (mostly in python).