
We're hiring! Come join the team and scale new heights with us! 🏔️
arcinstitute.org/jobs
@yusufroohani.bsky.social
Machine Learning & Systems Biology. ML Group Leader @arcinstitute. PhD @StanfordAILab http://www.yusufroohani.com

We're hiring! Come join the team and scale new heights with us! 🏔️
arcinstitute.org/jobs
scBaseCamp is released as part of the Arc Virtual Cell Atlas!
Great work by Nick Youngblut, Chris Carpenter, Alex Dobin, Dave Burke, @genophoria.bsky.social and team
📢Announcement: arcinstitute.org/news/news/ar...
🔗Data access: github.com/ArcInstitute...
📄Report: arcinstitute.org/manuscripts/...
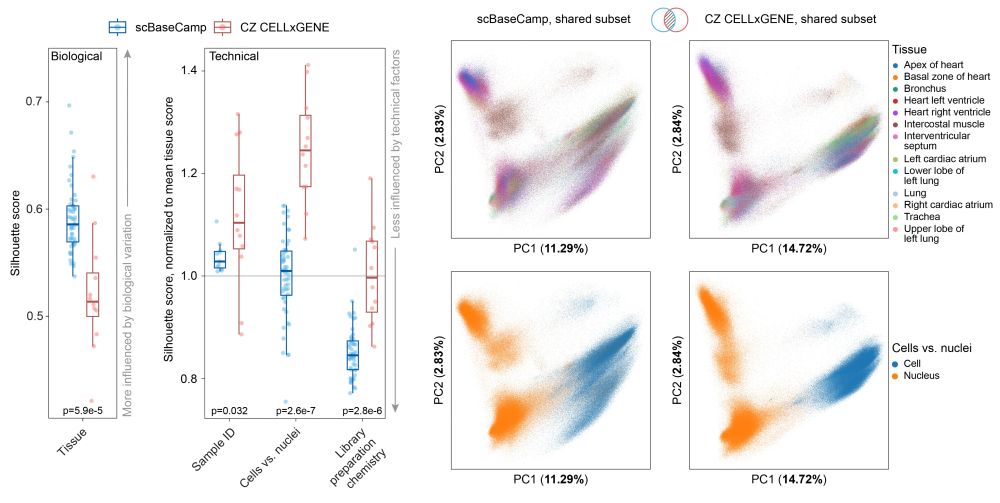
Uniform processing lowers technical variation between scBaseCamp datasets.
Technical factors such as library chemistry and suspension type (single-cell vs single-nucleus) exhibited comparable or lower silhouette scores than biologically meaningful categories like tissue type
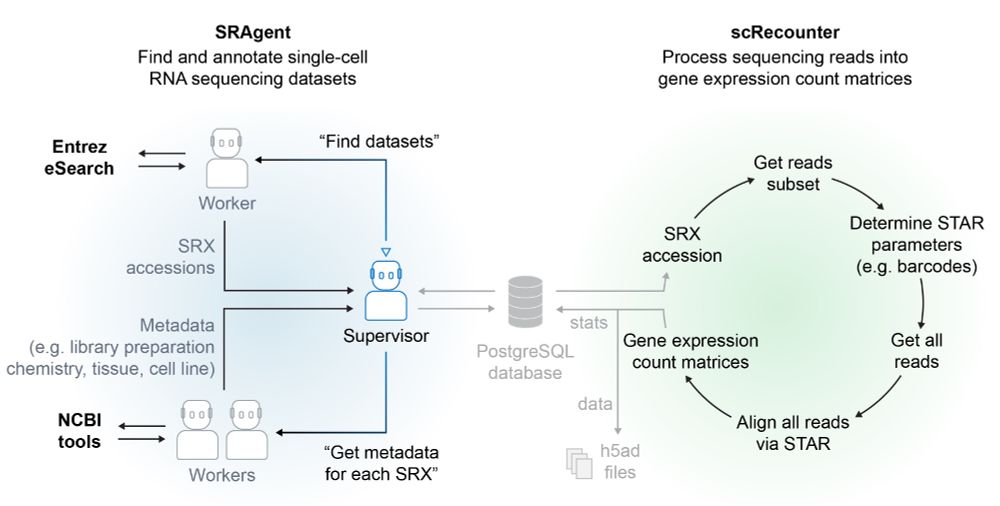
scBaseCamp is the first large biological data repository curated by an AI agent
We built a hierarchical agentic workflow (SRAgent) to automate discovery, metadata extraction & data processing
It is consistent, easily scalable and automatically updates when new data is available
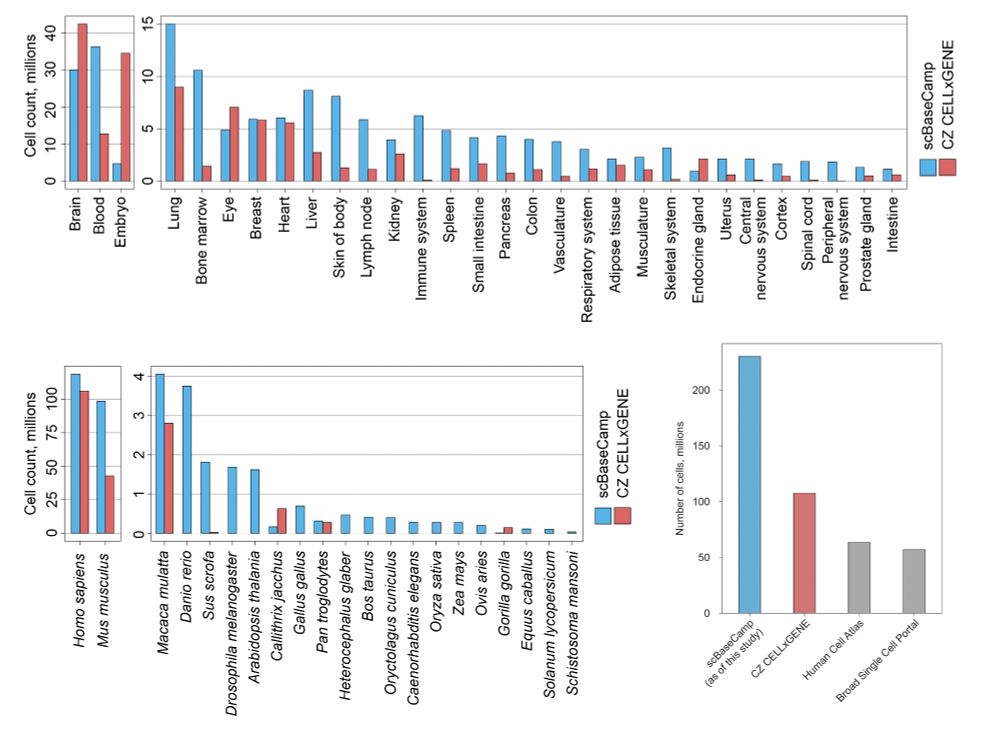
scBaseCamp was built by directly mining all publicly accessible 10X Genomics scRNAseq data from the Sequence Read Archive (SRA)
With over 230M cells drawn from 21 species and 72 tissues, scBaseCamp is significantly larger and more diverse than existing single-cell data repositories

At the @arcinstitute.org we are building AI models of cell state from the ground up, rethinking every step, from data generation to biologically relevant evaluation
Today we launch scBaseCamp, the largest public repository of single cell RNAseq data, uniformly processed from raw sequencing reads.
Why do you want to switch
04.12.2024 21:01 — 👍 0 🔁 0 💬 1 📌 0very easy to do this in Pycharm
04.12.2024 20:43 — 👍 1 🔁 0 💬 1 📌 0