Incredible work, wow! Massive congratulations to you and your team!!!
21.08.2025 06:50 —
👍 0
🔁 0
💬 0
📌 0
STING induces ZBP1-mediated necroptosis independently of TNFR1 and FADD - Nature
Nature - STING induces ZBP1-mediated necroptosis independently of TNFR1 and FADD
I am beyond excited and incredibly proud to share that our work is now out in @nature.com This discovery was made possible thanks to the first author Kostas kelepouras, my fantastic lab , and the unique research environment here in Cologne in collaboration with the Hospital bambino Gesu in Rome.
21.08.2025 06:12 —
👍 212
🔁 105
💬 2
📌 0
Congratulations Debora, fantastic news!
01.08.2025 13:57 —
👍 3
🔁 0
💬 0
📌 0
Next one is coming up, will contact you soon!
29.07.2025 12:13 —
👍 1
🔁 0
💬 0
📌 0
Thanks Karolin, all the credit to Gloria Fuentes @gloglita.bsky.social.
28.07.2025 18:46 —
👍 1
🔁 0
💬 0
📌 0
A great skeetorial on a new and much improved method for mapping transcription factor landscapes, with a beautiful image by our own Gloria Fuentes @gloglita.bsky.social from Life Science Editors @lifescienceeditors.bsky.social
28.07.2025 17:02 —
👍 7
🔁 2
💬 0
📌 0
🚀Big news from CRC 1678!
Robert Hänsel-Hertsch @epistrucstab.bsky.social and team unveil DynaTag—a single-cell method to map protein-DNA binding with stunning precision. Outshines ChIP-seq & CUT&RUN.
🧬Cancer research & gene regulation just got a major upgrade. #Genomics #DynaTag #PrecisionMedicine
28.07.2025 15:30 —
👍 3
🔁 1
💬 0
📌 0
🔬 DynaTag by CRC 1678 project leader Robert Hänsel-Hertsch @epistrucstab.bsky.social maps protein-DNA binding at single-cell level—boosting precision in cancer & gene research. #CRC1678 #DynaTag #UniCologne
28.07.2025 15:35 —
👍 5
🔁 1
💬 0
📌 0
Thank you Son for your invaluable help with the skeet! Your scientific illustrations look outstanding!
28.07.2025 11:52 —
👍 1
🔁 0
💬 1
📌 0
🧪New method for anyone interested in protein-DNA interactions and TF mapping, from the lab of @epistrucstab.bsky.social at @unicologne.bsky.social!👇
28.07.2025 11:44 —
👍 10
🔁 1
💬 1
📌 0
Highly recommended read for everyone interested in interactions of proteins and DNA: Impressive new method on single cell level introduced by @epistrucstab.bsky.social from @unicologne.bsky.social!
28.07.2025 10:41 —
👍 5
🔁 2
💬 0
📌 0
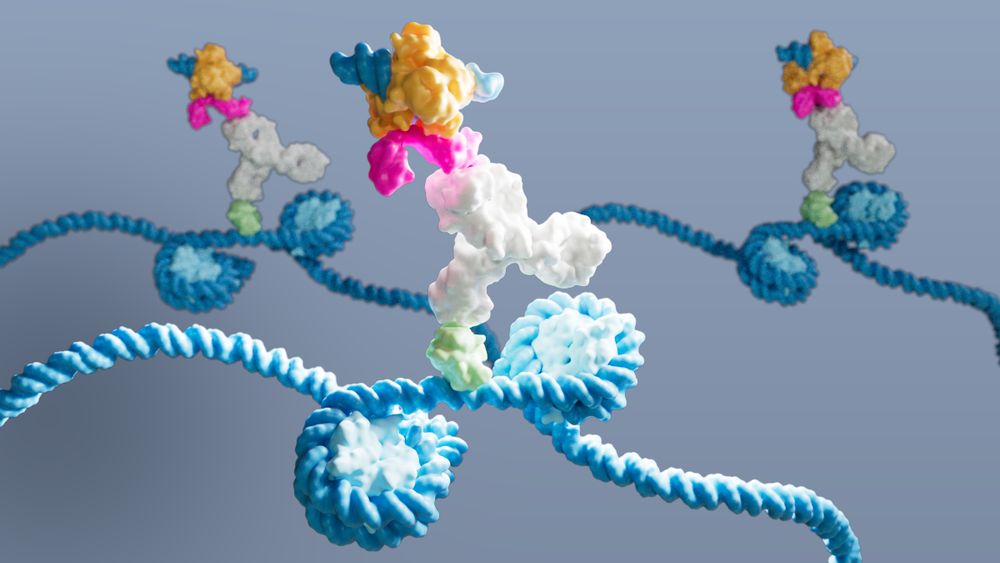
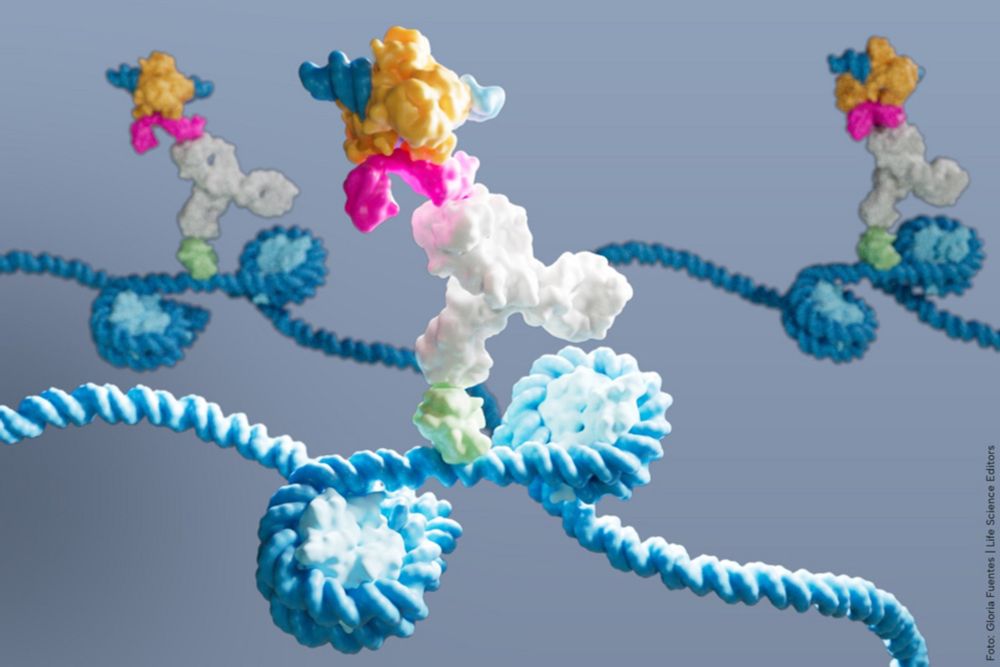
Image credit: @gloglita.bsky.social @lifescienceeditors.bsky.social captured DynaTag in action: a pA-Tn5 probe (multicoloured) binds an antibody (white), which binds p53 DNA-binding domain (green) on DNA (blue) within 2 nucleosomes
Image credit: @gloglita.bsky.social @lifescienceeditors.bsky.social captured DynaTag in action: a pA-Tn5 probe (multicoloured) binds an antibody (white), which binds p53 DNA-binding domain (green) on DNA (blue) within 2 nucleosomes
28.07.2025 09:10 —
👍 4
🔁 0
💬 0
📌 0
Thanks to former PhD student Pascal Hunold and lab members Giulia Pizzolato & Olivia van Ray for their amazing work. Grateful to @dtg-cologne.bsky.social, labs of Peifer, George & Thomas, funders #CMMC @dfg.de #sfb1399 @crc1678.bsky.social #FOR5504 #Fritz_Thyssen_Foundation (7/7)
28.07.2025 09:10 —
👍 4
🔁 0
💬 1
📌 0
The University of Cologne press release can be found here:👇
www.uni-koeln.de/universitaet...
www.uni-koeln.de/en/universit... (6/7)
28.07.2025 09:10 —
👍 2
🔁 0
💬 1
📌 0
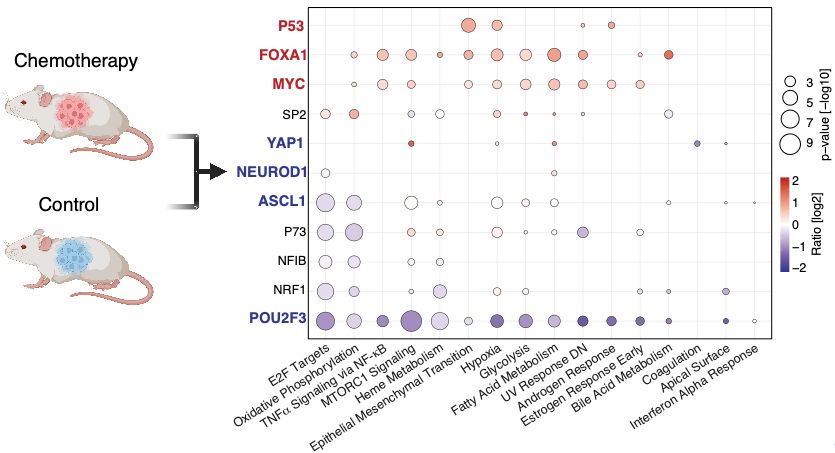
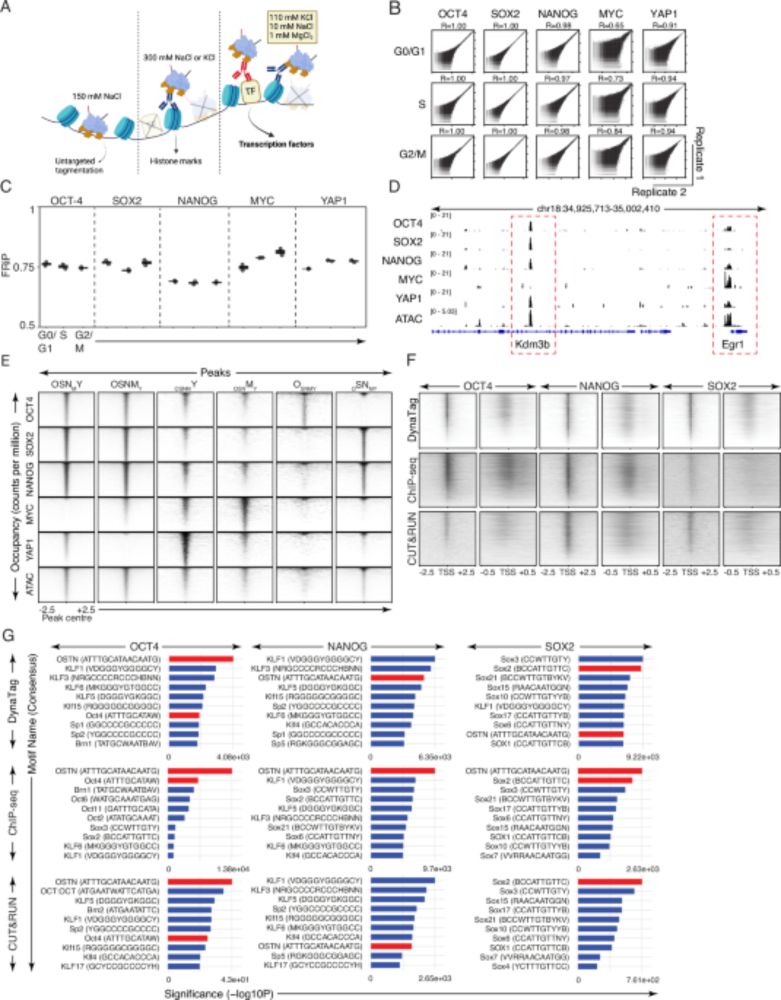
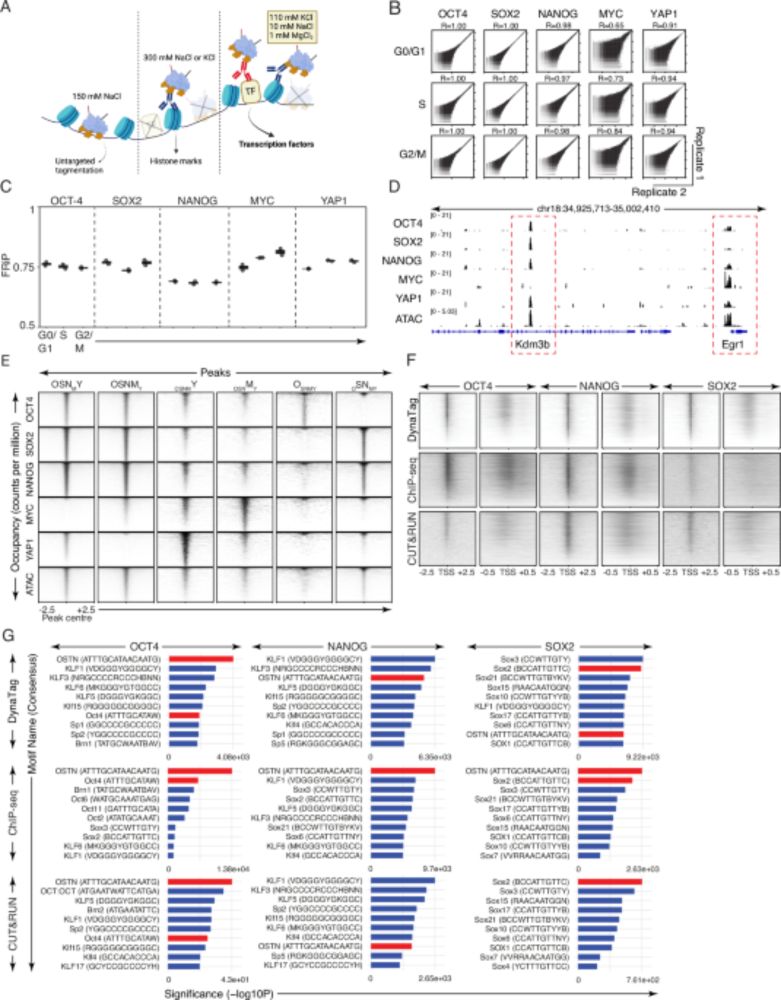
Adapted from Fig. 4 --> https://www.nature.com/articles/s41467-025-61797-9
🗝️ #DynaTag can reveal nuanced TF occupancy behaviour and shed light on transcriptional regulation in health and disease. In small cell lung cancer #SCLC PDX models, it uncovered surprising gain-of-function p53, FOXA1 & MYC activity post-chemotherapy – not for ASCL1, NEUROD1, POU2F3 or YAP1 (5/7)
28.07.2025 09:10 —
👍 3
🔁 0
💬 1
📌 0
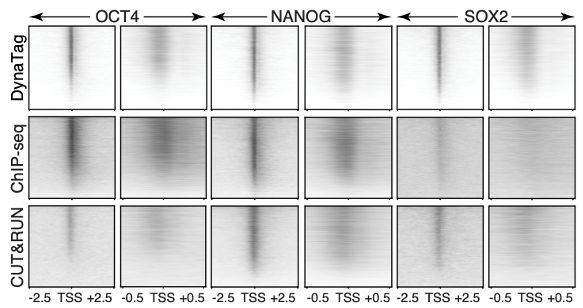
Fig.1f from https://www.nature.com/articles/s41467-025-61797-9
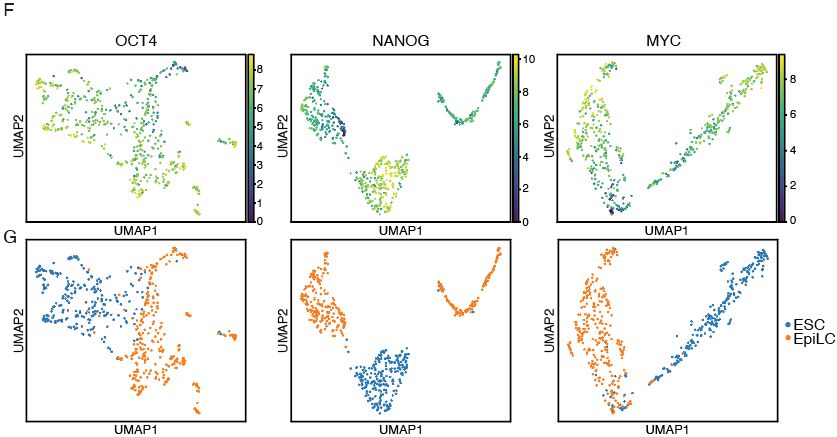
Fig.3f and g from https://www.nature.com/articles/s41467-025-61797-9
How is DynaTag better than other technologies?
⚡Preserves weak/transient TF binding without signal loss
⚡Better signal-to-noise & resolution
⚡Works for all TFs (high and low DNA-binding affinity) & histone marks
⚡Low-input needed, bulk or single-cell, scalable to multi-cellular systems (4/7)
28.07.2025 09:10 —
👍 4
🔁 0
💬 1
📌 0
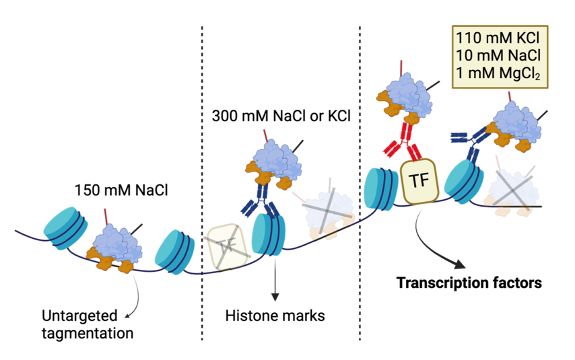
Fig.1a from https://www.nature.com/articles/s41467-025-61797-9
🤔 How to capture nuanced TF-DNA interactions?
💡 Enter #DynaTag: ‘Dynamic targets and Tagmentation’ captures transient TF-DNA interactions that standard CUT&Tag misses.
The trick? Sample prep under physiological salt stabilizes specific interactions without promoting untargeted tagmentation 🧂 (3/7)
28.07.2025 09:10 —
👍 5
🔁 0
💬 1
📌 0
❗TF occupancy mapping is still challenging. CUT&Tag was a breakthrough, but it has limits…
👉 Doesn’t work well for TFs that only transiently bind DNA
👉 These fleeting interactions are often lost during sample prep
That means we’re missing key regulatory events and it’s time for a new method (2/7)
28.07.2025 09:10 —
👍 4
🔁 0
💬 1
📌 0

Image credit: @gloglita.bsky.social @lifescienceeditors.bsky.social captured DynaTag in action: a pA-Tn5 probe (multicoloured) binds an antibody (white), which binds p53 DNA-binding domain (green) on DNA (blue) within 2 nucleosomes
🧪Move over CUT&Tag, there’s a new #TranscriptionFactor mapping method in town.
Our newly developed DynaTag is faster, cleaner, more sensitive than #ChIPseq, #CUT&RUN and #CUT&Tag.
🔗 Our @natcomms.nature.com paper: www.nature.com/articles/s41...
🧵Let’s break down what makes DynaTag so powerful (1/7)
28.07.2025 09:10 —
👍 94
🔁 30
💬 4
📌 4
Incredible!
29.06.2025 17:42 —
👍 1
🔁 0
💬 0
📌 0
This project is funded by our DFG @crc1678.bsky.social
27.06.2025 12:46 —
👍 1
🔁 0
💬 0
📌 0

Very proud of Dalya Hiluf being among the poster prize winners at the recent EMBO | EMBL Symposium 'The ageing genome'. Great stuff and congratulations!
27.06.2025 12:44 —
👍 3
🔁 0
💬 1
📌 1