I have more features planned to support my work, for instance multiple constructs in one library, combinations of sub-libraries and a region type expecting variants to a base sequence, but issues and pull-requests with other bugs and suggestions are very welcome too.
13.10.2025 14:35 — 👍 0 🔁 0 💬 1 📌 0
Simulated tests and benchmarks and our datasets suggest the tool is generally accurate and robust as well as pretty quick. Feels like a state that could be useful more widely so a good time to share, although I do expect more bugs to come out with wider usage!
13.10.2025 14:35 — 👍 0 🔁 0 💬 1 📌 0
Currently position in reads, flanking patterns and full alignment can be used to extract regions and extact matching, hamming distance and (bounded) Levenshtein used to compare to your library. This lets you nicely balance match accuracy with speed for your design.
13.10.2025 14:35 — 👍 2 🔁 0 💬 1 📌 0
It takes in (paired) fastq/a files, a JSON expected read structure, an optional expected combinations TSV and a strategy for extracting and matching variable regions and outputs count tables against your library.
13.10.2025 14:35 — 👍 0 🔁 0 💬 1 📌 0
MPRAbase a Massively Parallel Reporter Assay database
An international, peer-reviewed genome sciences journal featuring outstanding original research that offers novel insights into the biology of all organisms
MPRAbase (mprabase.ucsf.edu) , a customized database for massively parallel reporter assays (MPRAs) to easily find and download MPRA data. Amazing work by Jingjing Zhao, Fotis Baltoumas, Georgios Pavlopoulos, @vagar.bsky.social, ilias Georgakopoulos-Soares & others.
genome.cshlp.org/content/earl...
22.04.2025 20:51 — 👍 30 🔁 10 💬 0 📌 0
We're hiring to expand on the work to understand the human genome by engineering it!
lnkd.in/da-gitNc
10.02.2025 08:53 — 👍 12 🔁 8 💬 0 📌 0
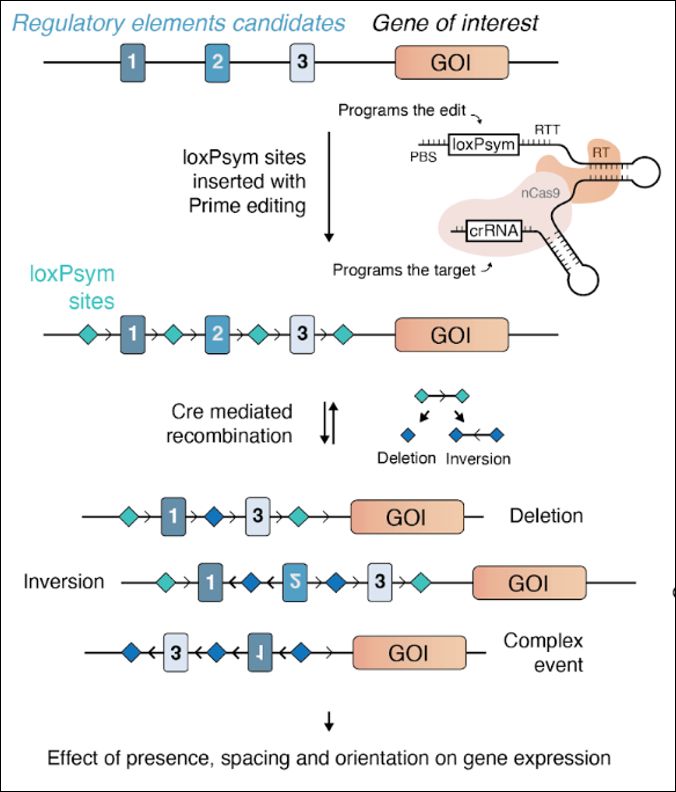
Enhancer scrambling strategy
We are happy to share our enhancer scramble story, a strategy to create hundreds of stochastic deletions, inversions, and duplications within mammalian gene regulatory regions and associate these new architectures with gene expression levels 🧵
www.biorxiv.org/content/10.1...
15.01.2025 20:32 — 👍 182 🔁 77 💬 3 📌 2
LinkedIn
This link will take you to a page that’s not on LinkedIn
Does my mutation have the same impact as yours? Population genetics 🤠 🥸 🤓 🤡 meets single cell CRISPRi ⚡ ! www.biorxiv.org/content/10.1... Led by Claudia Feng, Oliver Stegle, Britta Velten, @sangerinstitute.bsky.social .
02.12.2024 13:57 — 👍 58 🔁 24 💬 2 📌 3
Postdoc in Lars Velten's lab @crg.eu, working on gene regulation in haematopoiesis using ML techniques
https://jacobhepkema.github.io
Postdoc at UCL with James Reading. Previously at EMBL working with Wolfgang Huber. Biostats, R, cancer immunology
Ancient & military historian specializing in the Roman economy and military. PhD from UNC History. More impressive credential is that I have beaten both Dark Souls and Elden Ring.
Blogs at acoup.blog
We are a world leader in genomics research. We apply and explore genomic technologies to advance the understanding of biology and improve health 🧬
https://www.sanger.ac.uk/
Associate professor at ETH Zurich, studying the cellular consequences of genetic variation. Affiliated with the Swiss Institute of Bioinformatics and a part of the LOOP Zurich.
Human evolutionary genetics, University of Cambridge; Darwin College. 🇮🇪
Scientist developing AI for oncology. Division head at the German Cancer Research Centre DKFZ. Prof at the University of Heidelberg, Germany. Previously at EMBL-EBI and Wellcome Sanger Institute. Alumnus of ETH Zurich.
Group Leader in Human Genetics, Wellcome Sanger Institute
Executive Director EMBL. I have an insatiable love of biology. Consultant to ONT and Cantata (Dovetail)
Solve Biology
Head of Generative Genomics, Wellcome Sanger Institute, Cambridge, UK
Systems + Synthetic Biology, CRG, Barcelona
http://barcelonacollaboratorium.com http://allox.bio https://www.sanger.ac.uk/programme/generative-and-synthetic-genomics/
Wellcome Trust Clinical PhD Fellow in the Garnett Lab @sangerinstitute.bsky.social. Interested in cancer plasticity, multiplexed assays, ML, & comp bio.
https://ttszen.com/
Probabilistic machine learning to address questions in evolution and health #EvolutionaryMedicine. PI at the Centre for Genomic Regulation, co-leading a group with Mafalda Dias. Previously Harvard.
Scientist at University of Edinburgh. Director of MRC Human Genetics Unit but posting in my own capacity.
Scientist. Group leader at the Sanger Institute, Cambridge UK. Somatic mutation and selection in normal tissues, cancer and ageing.
Head of Human Genetics & Senior Group Leader @ Wellcome Sanger Institute. Statistical geneticist working on complex diseases #ibd #firstgen.
Researcher @bayraktar_lab @teichlab @steglelab.bsky.social @sangerinstitute.bsky.social | Using models & AI to study cells, cell circuits & brains 🧠 | #SingleCell+spatial | 🌍+🇺🇦
Cell Scientist, CRISPRist, @ Wellcome Sanger Institute,
stem cell nomad hunting new drug targets in Cancer,
Cambridge, UK
Target discovery | Computational Biology | Human Disease Genetics | ML | Cloud computing - @opentargets.org
- @ebi.embl.org
Statistician, Computational Biologist, R |> Bioconductor
https://www.huber.embl.de
Textbook: Modern Statistics for Modern Biology https://www.huber.embl.de/msmb/ (with @sherlockpholmes.bsky.social)
Cancer Research UK Career Development Fellow at the Wellcome Sanger Institute - gene editing - drug resistance - cancer


