De-novo promoters emerge more readily from random DNA than from genomic DNA
Promoters are DNA sequences that help to initiate transcription. Point mutations can create de-novo promoters, which can consequently transcribe inactive genes or create novel transcripts. We know lit...
Excited / nervous to share the “magnum opus” of my postdoc in Andreas Wagner’s lab!
"De-novo promoters emerge more readily from random DNA than from genomic DNA"
This project is the accumulation of 4 years of work, and lays the foundation for my future group. In short, we… (1/4)
28.08.2025 06:37 — 👍 167 🔁 59 💬 4 📌 1

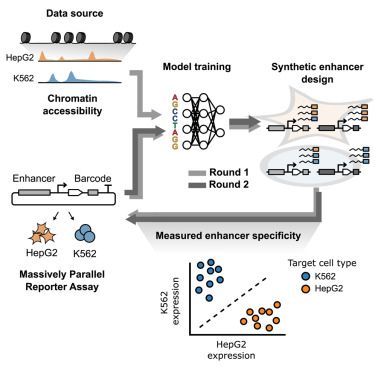
Activity of most genes is controlled by multiple enhancers, but is there activation coordinated? We leveraged Nanopore to identify a specific set of elements that are simultaneously accessible on the same DNA molecules and are coordinated in their activation. www.biorxiv.org/content/10.1...
18.08.2025 12:23 — 👍 95 🔁 39 💬 2 📌 2

Range extender mediates long-distance enhancer activity - Nature
The REX element is associated with long-range enhancer–promoter interactions.
Our paper describing the Range Extender element which is required and sufficient for long-range enhancer activation at the Shh locus is now available at @nature.com. Congrats to @gracebower.bsky.social who led the study. Below is a brief summary of the main findings www.nature.com/articles/s41... 1/
02.07.2025 16:17 — 👍 185 🔁 90 💬 10 📌 9
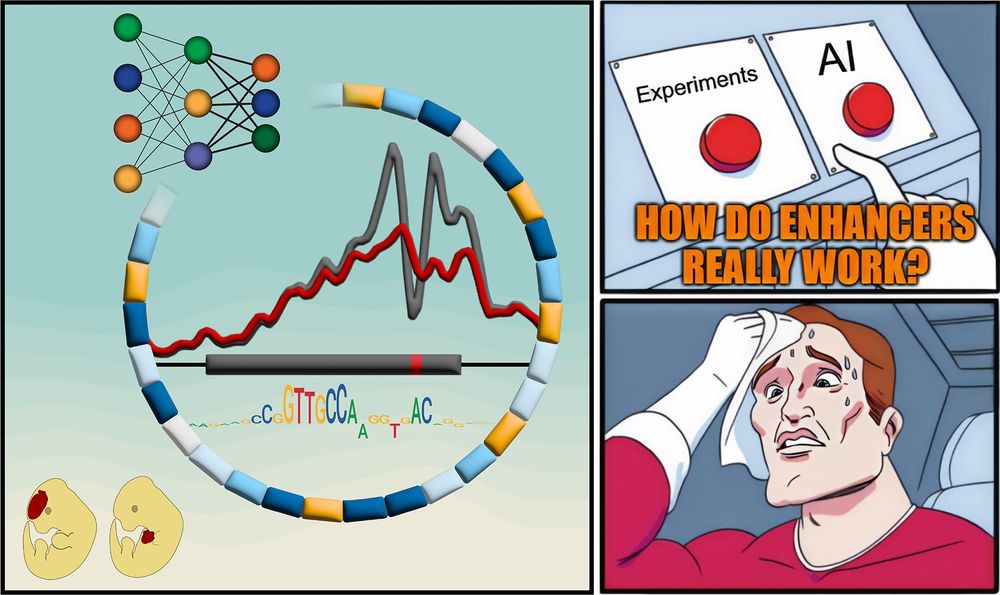
A meme-style comic panel with three parts. Left: A stylized enhancer with a mutation, surrounded by colored blocks representing functional motifs, a neural network diagram, chromatin accessibility signal traces, and a sequence motif. Two cartoon mouse embryos below show different LacZ reporter activity patterns. Top right: A hand hovers anxiously between two red buttons labeled “Experiments” and “AI,” with the caption “HOW DO ENHANCERS REALLY WORK?” Bottom right: A sweating superhero wipes his forehead, looking stressed about the difficult choice.
Textbooks: “Enhancers are just a bunch of TFBSs”
But how do they REALLY work?
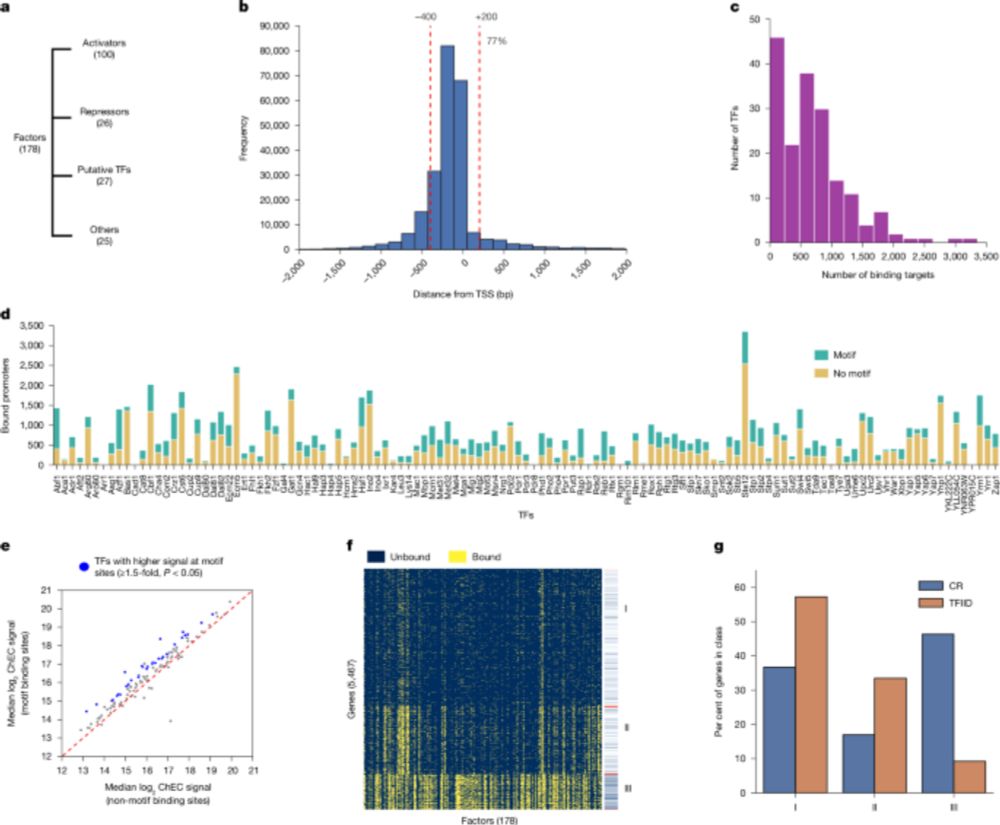
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22
18.06.2025 17:55 — 👍 165 🔁 80 💬 2 📌 5

Conservation of regulatory elements with highly diverged sequences across large evolutionary distances
Nature Genetics - Combining functional genomic data from mouse and chicken with a synteny-based strategy identifies positionally conserved cis-regulatory elements in the absence of direct sequence...
How to find Evolutionary Conserved Enhancers in 2025? 🐣-🐭
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
27.05.2025 12:19 — 👍 245 🔁 110 💬 7 📌 9
Our latest work now online in Cell:
Rewriting regulatory DNA to dissect and reprogram gene expression
Our new method (Variant-EFFECTS) uses high-throughput prime editing + flow sorting + sequencing to precisely measure effects of noncoding variants on gene expression
Thread 👇
17.04.2025 18:26 — 👍 117 🔁 28 💬 1 📌 2
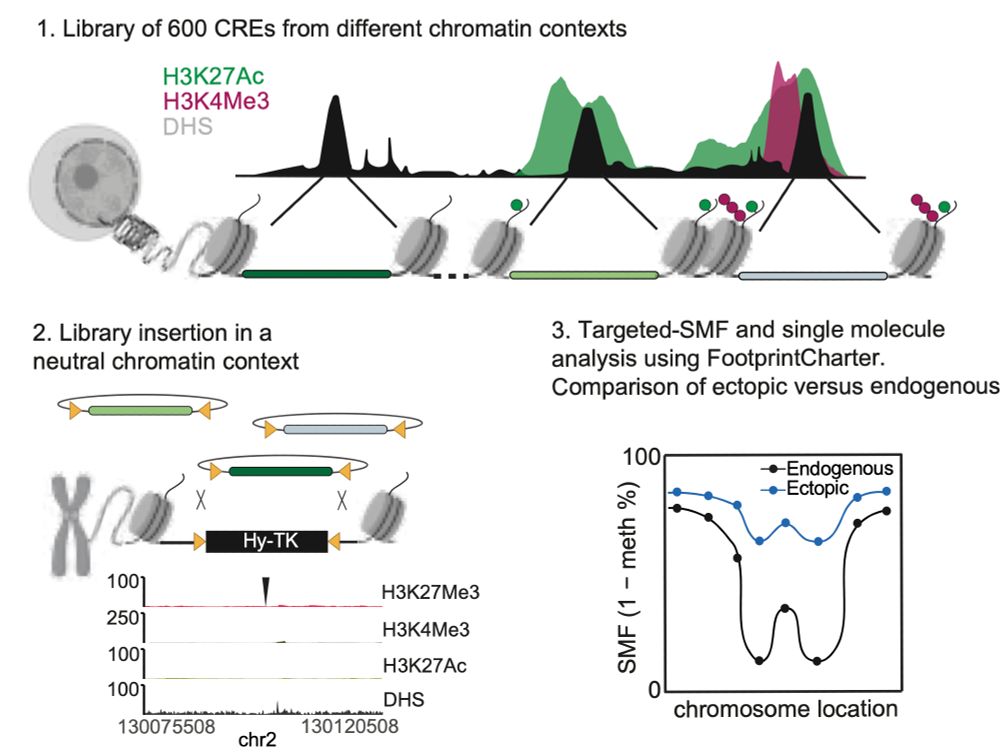
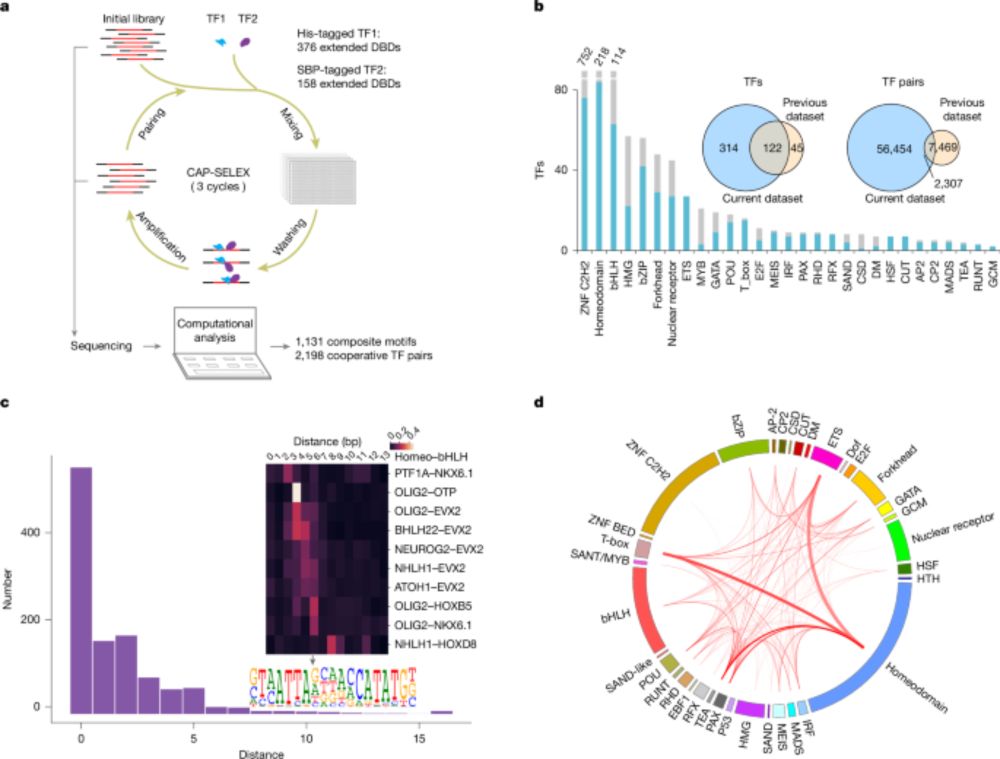
Happy to share the latest story from @arnaudkr.bsky.social's lab @embl.org! With @guidobarzaghi.bsky.social, we used Single Molecule Footprinting to quantify how often chromatin is accessible at enhancers after TF and chromatin environment changes! Check our preprint bit.ly/3XQMFxN + thread ⬇️ 1/11
08.04.2025 13:51 — 👍 77 🔁 33 💬 4 📌 2
Our new preprint is out! Want to better visualize what your sequence-to-function profile learned? Here is PISA. It also comes in a new BPNet package, which can be used to train many genomics data sets, including MNase-seq data.
08.04.2025 13:31 — 👍 26 🔁 8 💬 1 📌 0
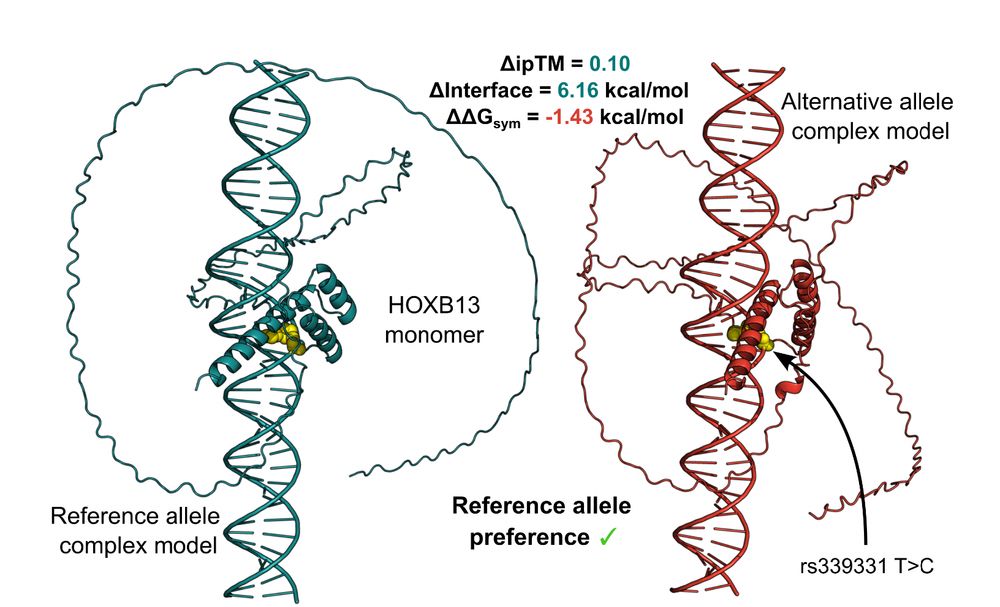
Unpicking non-coding genetic variation: Structure-guided modelling holds promise for evaluating how single nucleotide variants affect transcription factor binding. www.biorxiv.org/content/10.1.... @uoe-igc.bsky.social
21.03.2025 10:22 — 👍 16 🔁 8 💬 0 📌 0
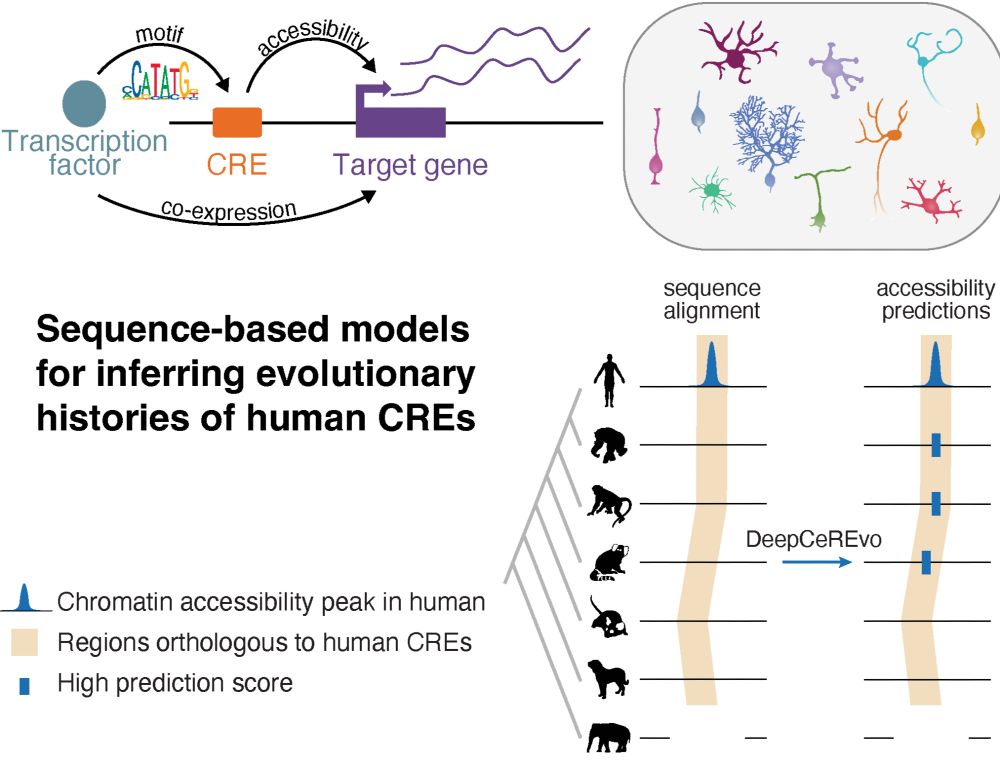
How does gene regulation shape brain evolution? Our new preprint dives into this question in the context of mammalian cerebellum development! rb.gy/dbcxjz
Led by @ioansarr.bsky.social, @marisepp.bsky.social and @tyamadat.bsky.social, in collaboration with @steinaerts.bsky.social
16.03.2025 10:31 — 👍 188 🔁 69 💬 4 📌 5
Just very happy to have our paper out today! A big thanks to all our co-authors, and to Nikolai and @steinaerts.bsky.social for the teamwork over the past years. If you are interested in using our models for cross-species enhancer studies, check out crested.readthedocs.io/en/stable/mo... 🙂
14.02.2025 10:07 — 👍 53 🔁 25 💬 3 📌 3
Recently, we've been playing around with using Ledidi to design "affinity catalogs" that exhibit a broad range of activities, rather than just the "most" of a desired activity.
Here are three example catalogs designed for GATA2 binding, chromatin accessibility, and transcription initiation.
12.02.2025 12:17 — 👍 19 🔁 7 💬 2 📌 0
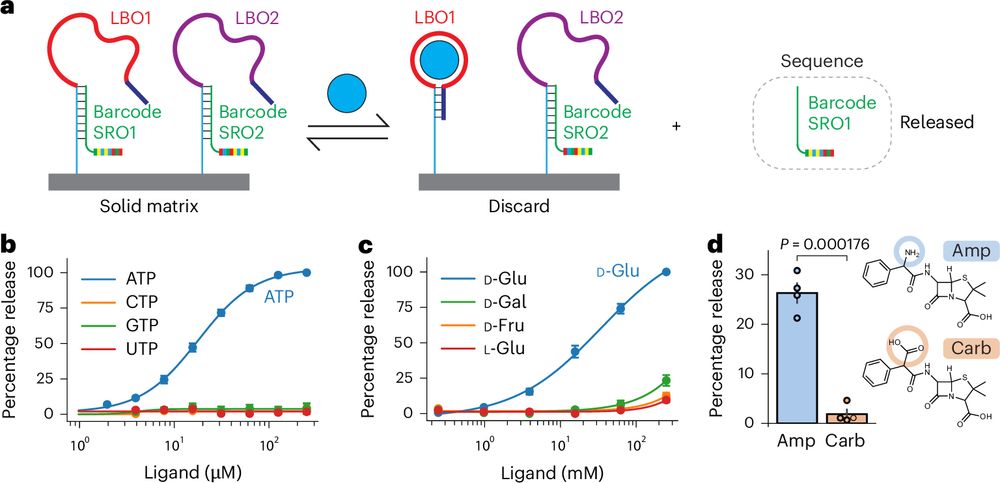
Quantifying metabolites using structure-switching aptamers coupled to DNA sequencing
Nature Biotechnology - Metabolites can be quantified using a combination of aptamers and DNA barcodes.
Delighted to share our latest paper describing a method to read the levels of hundreds of metabolites or drugs in parallel using DNA sequencing. This method, which we call ‘smol-seq’ (Small MOLecule sequencing), harnesses the power of DNA sequencing for metabolite detection:
rdcu.be/d8xLv (1/6)
04.02.2025 13:54 — 👍 159 🔁 62 💬 11 📌 11
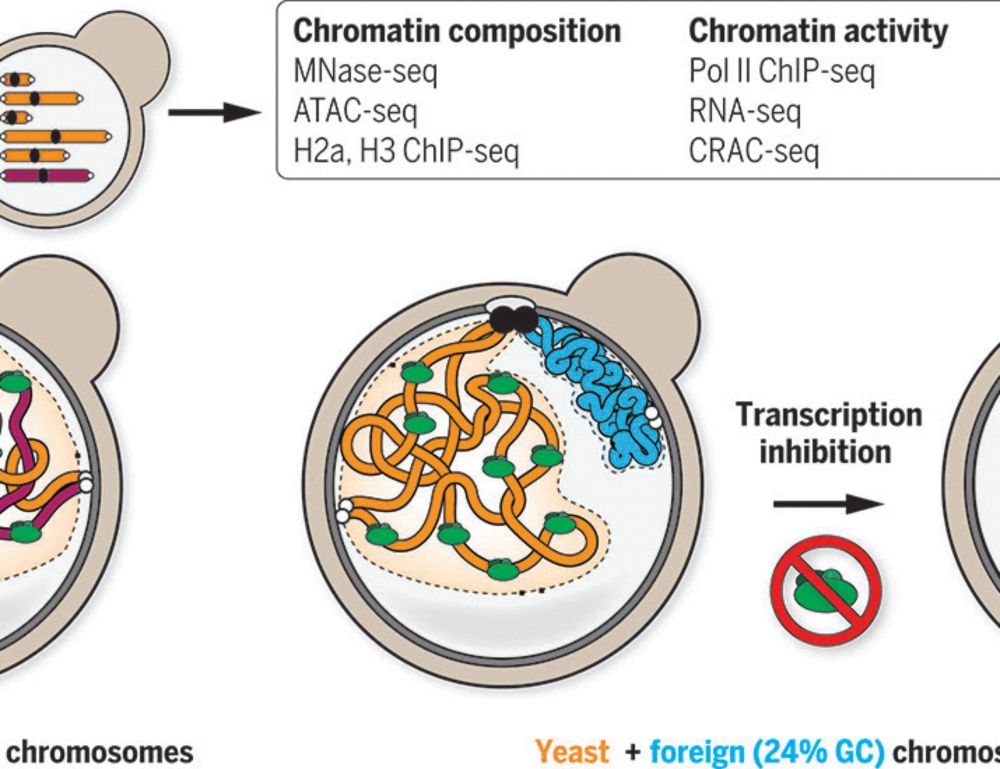
0/ Essential reading for anyone training or using sequence-function models trained on genomic sequences! 🚨 In our new preprint, we explore the ways homology within genomes can cause leakage when training sequence-based models and ways to prevent it
27.01.2025 23:04 — 👍 26 🔁 12 💬 1 📌 3
Looks like a very useful method for deeper chromatin accessibility analysis!
23.01.2025 07:34 — 👍 1 🔁 0 💬 0 📌 0
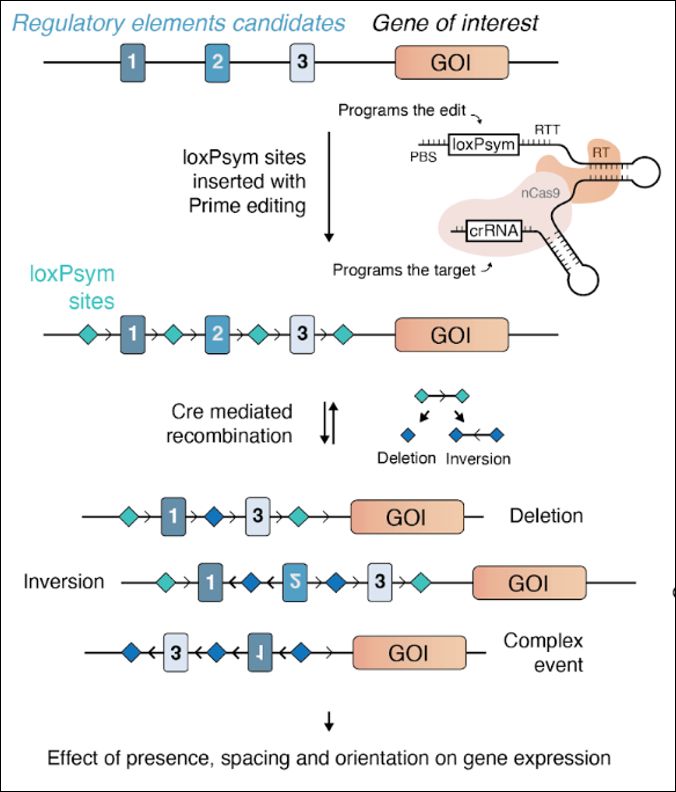
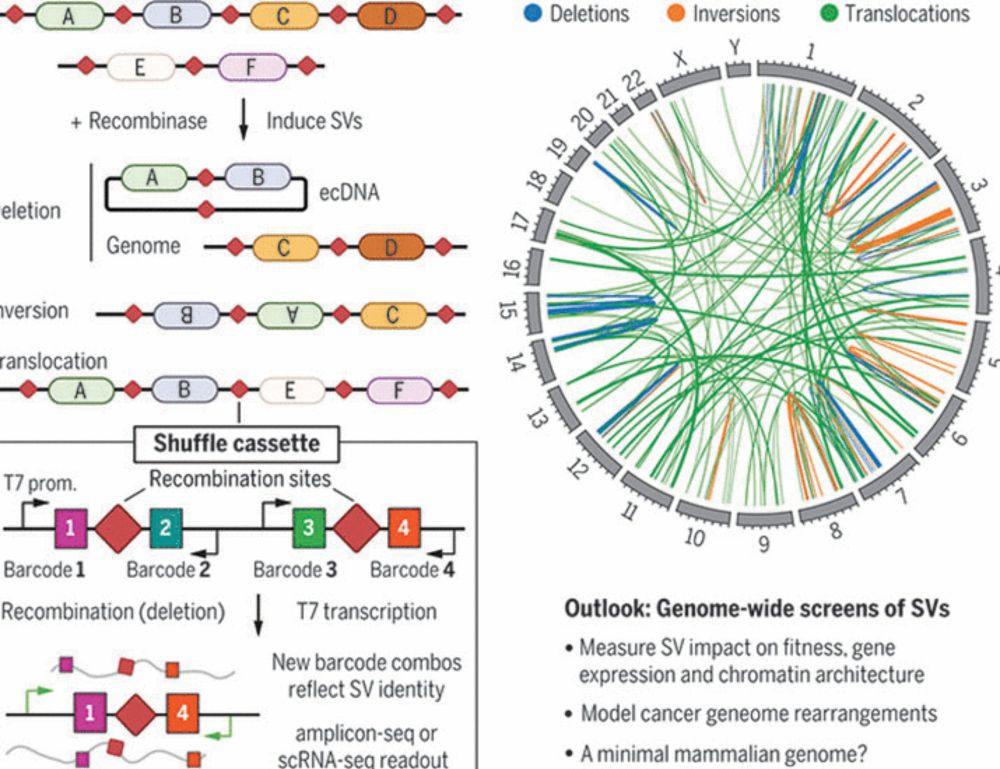
Enhancer scrambling strategy
We are happy to share our enhancer scramble story, a strategy to create hundreds of stochastic deletions, inversions, and duplications within mammalian gene regulatory regions and associate these new architectures with gene expression levels 🧵
www.biorxiv.org/content/10.1...
15.01.2025 20:32 — 👍 182 🔁 77 💬 3 📌 2
This is really cool - quite precise control! I especially like the collision avoidance system (I'm imagining parking beeps with each exclamation mark)
18.12.2024 09:17 — 👍 1 🔁 0 💬 0 📌 0
Congratulations Cansu!!
13.12.2024 12:17 — 👍 0 🔁 0 💬 1 📌 0
Really cool work!
03.12.2024 17:01 — 👍 2 🔁 0 💬 0 📌 0
Redirecting
By definition, enhancers can activate from a distance. But with increased distance between enhancer and promoter, the activation drops. To study this systematically, we build a synthetic locus: www.cell.com/molecular-ce... 1/12
02.12.2024 16:44 — 👍 226 🔁 83 💬 14 📌 11
LinkedIn
This link will take you to a page that’s not on LinkedIn
Does my mutation have the same impact as yours? Population genetics 🤠 🥸 🤓 🤡 meets single cell CRISPRi ⚡ ! www.biorxiv.org/content/10.1... Led by Claudia Feng, Oliver Stegle, Britta Velten, @sangerinstitute.bsky.social .
02.12.2024 13:57 — 👍 58 🔁 24 💬 2 📌 3
Group leader CRG; Associate Faculty ToL Sanger Institute.
Genome regulation, chromatin, cell types, and evolution. https://www.sebepedroslab.org
chromatin, transcription, lineage tracing & single cells
Opinions @WSJ.com & @Science.org. Author of Life Without Genes @harpercollins.bsky.social & On The Future of Species bloomsburybooksuk.bsky.social (12 February 2026). Founder genyro.com. Exploring the future of bioconvergence and generative biology.
Scientist at IMP in Vienna. Excited about gene expression regulation and its encoding in our genomes - enhancers, transcription factors, co-factors, silencers, AI.
We study transcriptional regulation and chromosome folding using an interdisciplinary approach combining wet- and dry-lab methods.
https://giorgettilab.org
@fmiscience.bsky.social
We are a research lab at the Netherlands Cancer Institute. We develop and apply new genomics tools to study genome biology and gene regulation.
Postdoc in Andreas Wagner's lab. I'm currently obsessing over how new promoters and enhancers emerge de novo. Also dogs 🐶 and triathlon 🏊 🚲 🏃. He/him 🏳🌈.
timothyfuqua.com
Scientist and medical doctor. Gene regulation, DNA sequence models, transcription factor binding. Doing a PhD in computational biology at @molgen.mpg.de.
tries to make microscopes smarter · bioimage analysis, optogenetics, ml, 3d printing, open science · phd student in cellular signalling dynamics @PertzLab
Postdoctoral Fellow, Genome Biology Unit, EMBL Heidelberg
Computational Pathology Biomarker Lead at AstraZeneca | PhD
Likes immune cells, single-cell technologies, and home made pizza
Postdoc at UCL with James Reading. Previously at EMBL working with Wolfgang Huber. Biostats, R, cancer immunology
Engineering cells to understand their decisions
ESPOD Fellow @ebi.embl.org & @sangerinstitute.bsky.social
(Saez-Rodriguez & Parts)
lingering scientist @crick.ac.uk (Briscoe)
Baylor College of Medicine
Ecological and Evolutionary Developmental Biologist on craniofacial tissues in reptile models 🐍🦎🐢🐊
Research Director @helsinki-biotech.bsky.social (https://www.helsinki.fi/en/researchgroups/vertebrate-evolution-development-and-regeneration)
Postdoc @sangerinstitute.bsky.social | Lover of all things single-cell ‘omic, common complex disease and genetics. Anderson lab - http://andersonlab.info
PhD student @sangerinstitute
Postdoctoral researcher at @anshulkundaje.bsky.social Machine learning #ML, gene regulatory networks #GRN, single cell and spatial #omics.
Previously at @saezlab.bsky.social
Developer of https://decoupler.readthedocs.io/