
Human organoid tumor transplantation identifies functional glioblastoma-microenvironment communication mediated by PTPRZ1
01.02.2026 00:23 — 👍 1 🔁 1 💬 0 📌 0@martyyang.bsky.social
Postdoc in Bhaduri Lab @ UCLA

Human organoid tumor transplantation identifies functional glioblastoma-microenvironment communication mediated by PTPRZ1
01.02.2026 00:23 — 👍 1 🔁 1 💬 0 📌 0
After a long review process, I'm excited that our paper is finally in print: www.cell.com/cell/fulltex...
TL;DR: We use CRISPR screens in iPSC-derived neurons to find a new tau E3 ligase and a relationship between oxidative stress, the proteasome, and tau proteolytic fragments.
More below 👇

1/ Our new study, led by Jingwen Ding, examines the role of transcription factors during human neurogenesis to identify gene regulatory networks influencing cell fate, maturation, and subtype specification
www.nature.com/articles/s41...
Here is a copy of last year's Twitter thread explaining our preprint - jump to (21) for the new stuff 👀
Synergy between cis-regulatory elements can render cohesin dispensable for distal enhancer function
now revised and journal accepted at www.science.org/doi/10.1126/...
🧵👇

Our new manuscript, led by Emily Corrigan, examines inhibitory neuron diversity across approximately 160 million years of evolutionary divergence, as part of BRAIN Initiative Cell Atlas Network (BICAN) developing brain atlas package: www.nature.com/articles/s41...
07.11.2025 18:06 — 👍 60 🔁 22 💬 2 📌 1
Very pleased to share this work from my time as a graduate student with the Greenberg lab. We investigate the molecular function of ZMYND11, a tumor suppressor and chromatin reader which is also a cause of syndromic intellectual disability. Preprint available now: www.biorxiv.org/content/10.1...
04.11.2025 23:37 — 👍 6 🔁 4 💬 1 📌 1🧠🌟🐭 Excited to share some of my postdoc work on the evolution of dexterity!
We compared deer mice evolved in forest vs prairie habitats. We found that forest mice have:
(1) more corticospinal neurons (CSNs)
(2) better hand dexterity
(3) more dexterous climbing, which is linked to CSN number🧵
The first (of hopefully many) reports to come from our collaboration with @hjp.bsky.social
We present a new type of cell fitness assay that allows you to both quantify and explain differences across human donors in cell proliferation and sensitivity to environmental toxicants.

Today I am so pleased to present our work on how chromatin remodelers affect mesoscale chromatin organization.
www.science.org/doi/10.1126/...
Super excited to get this out. This collab started a few years ago and is the first paper from it. Here, with experimental and computational approaches we:
1. establish that cell villages can be just as accurate (one might argue more accurate!) than arrayed-based designs
bsky.app/profile/bior...
Dissecting Gene Regulatory Networks Governing Human Cortical Cell Fate https://www.biorxiv.org/content/10.1101/2025.09.23.678137v1
24.09.2025 07:31 — 👍 3 🔁 1 💬 0 📌 0
Some (+)ve news to lighten another heavy weekend: our latest preprint (c/o Mattiroli + Ramani labs) is up!
www.biorxiv.org/content/10.1...
A tour-de-force by 1st authors Bruna Eckhardt & @palindromephd.bsky.social, focusing on chromatin replication. RTs welcome; tweetorial in 3,2...(1/n)
I'm really happy to announce that my two main postdoctoral works have been published in the same issue of Developmental Cell! First, the final form of the mouse cortical organoid protocol (tinyurl.com/bddfmx8n), which I recently presented at Development presents (tinyurl.com/36udcpme) and…
27.08.2025 16:25 — 👍 35 🔁 12 💬 8 📌 0A mouse organoid platform for modeling cerebral cortex development and cis-regulatory evolution in vitro: Developmental Cell www.cell.com/developmenta...
27.08.2025 15:09 — 👍 12 🔁 5 💬 0 📌 2This is not a HiC map! Ever wondered if multiple enhancers get activated simultaneously? We measured chromatin accessibility on thousands of molecules by nanopore to create genome-wide co-accessibility maps. Proud of @mathias-boulanger.bsky.social @kasitc.bsky.social Biology in the thread👇
18.08.2025 13:00 — 👍 113 🔁 34 💬 10 📌 0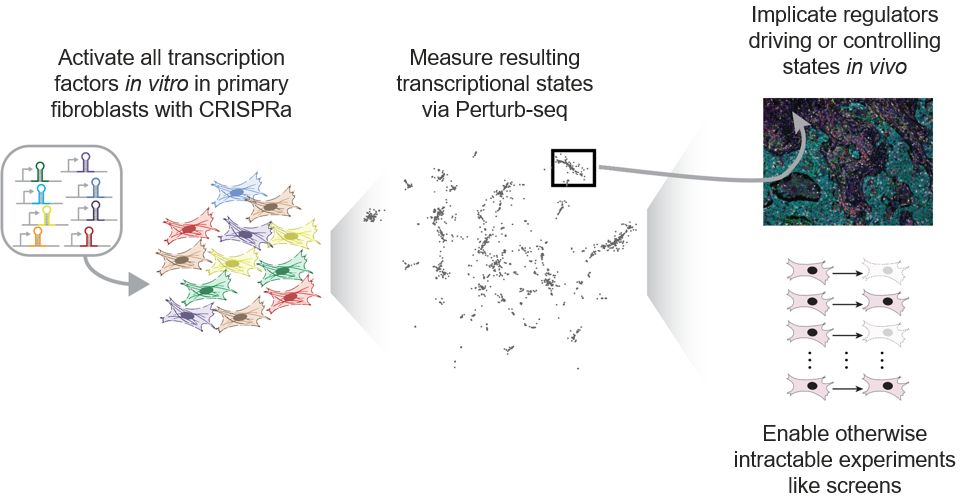
Our paper is now out in final form at Nature Genetics! For those who missed the preprint, we used large-scale Perturb-seq targeting transcription factors to push primary fibroblasts into diverse transcriptional states, including those observed in cell atlas studies.
06.08.2025 15:14 — 👍 74 🔁 29 💬 3 📌 1The Anne West lab at Duke is hiring a postdoc to work on nuclear cell biology in neurogenesis. Please help me spread the word! Applications to west@neuro.duke.edu.
27.07.2025 20:27 — 👍 58 🔁 51 💬 1 📌 2Two parallel lineage-committed progenitors contribute to the developing brain https://www.biorxiv.org/content/10.1101/2025.07.02.662771v1
03.07.2025 12:31 — 👍 2 🔁 1 💬 0 📌 0🧵Excited to share our new preprint introducing iHOTT - an autologous tumor-immune co-culture model that captures patient-specific responses in #Glioblastoma
💥Now on
@biorxivpreprint
: biorxiv.org/content/10.1...
Led by Dr. Shivani Baisiwala, Neurosurgery Resident in the lab

Excited to share our latest manuscript, a collaboration with @justinperryphd.bsky.social led by Weill Cornell Neuroscience PhD student @annbaako.bsky.social. Ann developed a new, improved mouse PSC model for studying tissue-resident macrophage differentiation.
www.biorxiv.org/content/10.1...
It’s out! The first paper from my postdoc – and first from the @bhadurilab.bsky.social – is now live @natneuro.nature.com . 🧠✨
Using a new meta-atlas generation strategy, we identified functional gene networks that more fully explain how cell types are formed in the human cortex. (1/13)

Preprint from @bhadurilab.bsky.social: Thalamic NRXN1-Mediated Input to Human Cortical Progenitors Drives Upper Layer Neurogenesis (www.biorxiv.org/content/10.1...)
27.04.2025 16:32 — 👍 1 🔁 0 💬 0 📌 0
What contributes to human specific neural specification? Check out these two wonderful preprints from a wonderful scientist Ava carter! 1. www.biorxiv.org/content/10.1...; 2. www.biorxiv.org/content/10.1...
Stay tuned! third preprint is on its way!

Delighted to share our most recent preprint led by the incredible @MilJessenya and @Jalbsoto! In this study, we generated a metabolomic atlas of human cortical development and discovered that the pentose phosphate pathway regulates radial glia cell fate. www.biorxiv.org/content/10.1...
13.03.2025 04:36 — 👍 16 🔁 6 💬 1 📌 1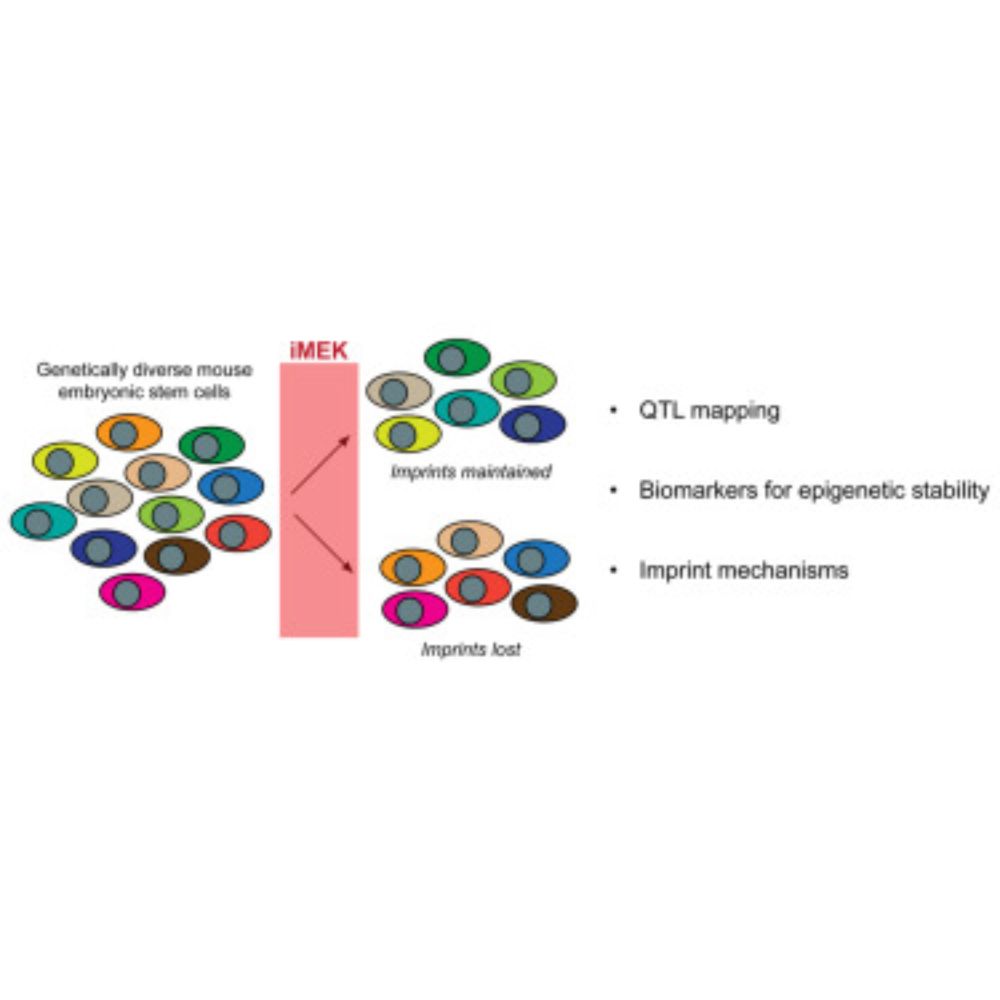
Our collaborative project with Matthias Stadtfeld and Jason Mezey labs has now been published. We examined how genetic variation impacts regulation of DNA methylation at the control regions of imprinted genes in naive pluripotent stem cells.
www.cell.com/stem-cell-re...

⚡Next #FragileNucleosome meeting is less than a week away! Three amazing ECR scientists, 🔬@anajaneva.bsky.social, 🧠@anastasiahains.bsky.social, & 🧬@martyyang.bsky.social will discuss their research next Wed!
🗓️Don't forget to register:
us06web.zoom.us/webinar/regi...
Pervasive and programmed nucleosome distortion patterns on single mammalian chromatin fibers https://www.biorxiv.org/content/10.1101/2025.01.17.633622v1
22.01.2025 18:39 — 👍 4 🔁 2 💬 0 📌 0Excited to share the second story from my post-doc in pre-print form! Many thanks to @vram142.bsky.social, lab members, and collaborators for contributions! In brief, we examine heterogeneity in (sub)nucleosome structure from in vivo long-read m6dA footprinting data. See Vijay's thread for details.
24.01.2025 01:14 — 👍 8 🔁 4 💬 0 📌 0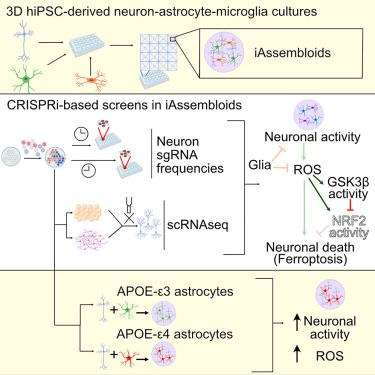
Our latest paper, led by Emmy Li, is out on Neuron today:
CRISPRi-based screens in iAssembloids to elucidate neuron-glia interactions
www.cell.com/neuron/fullt...