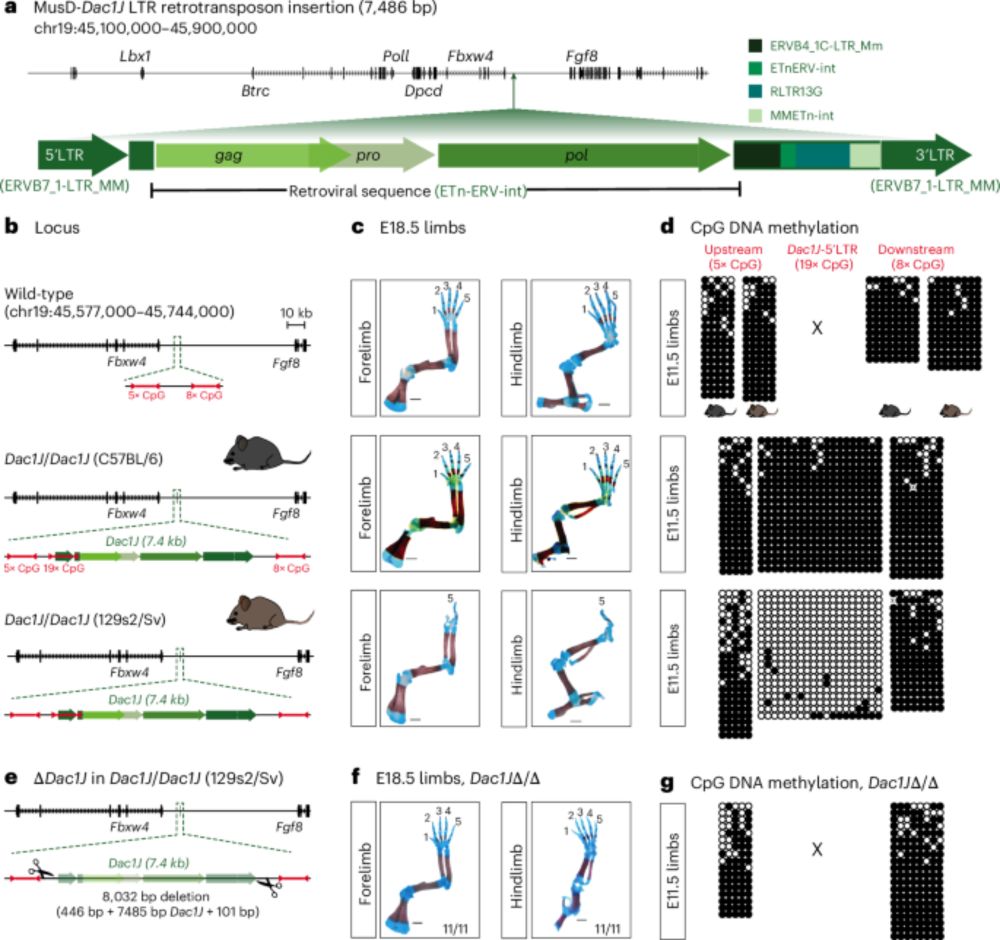
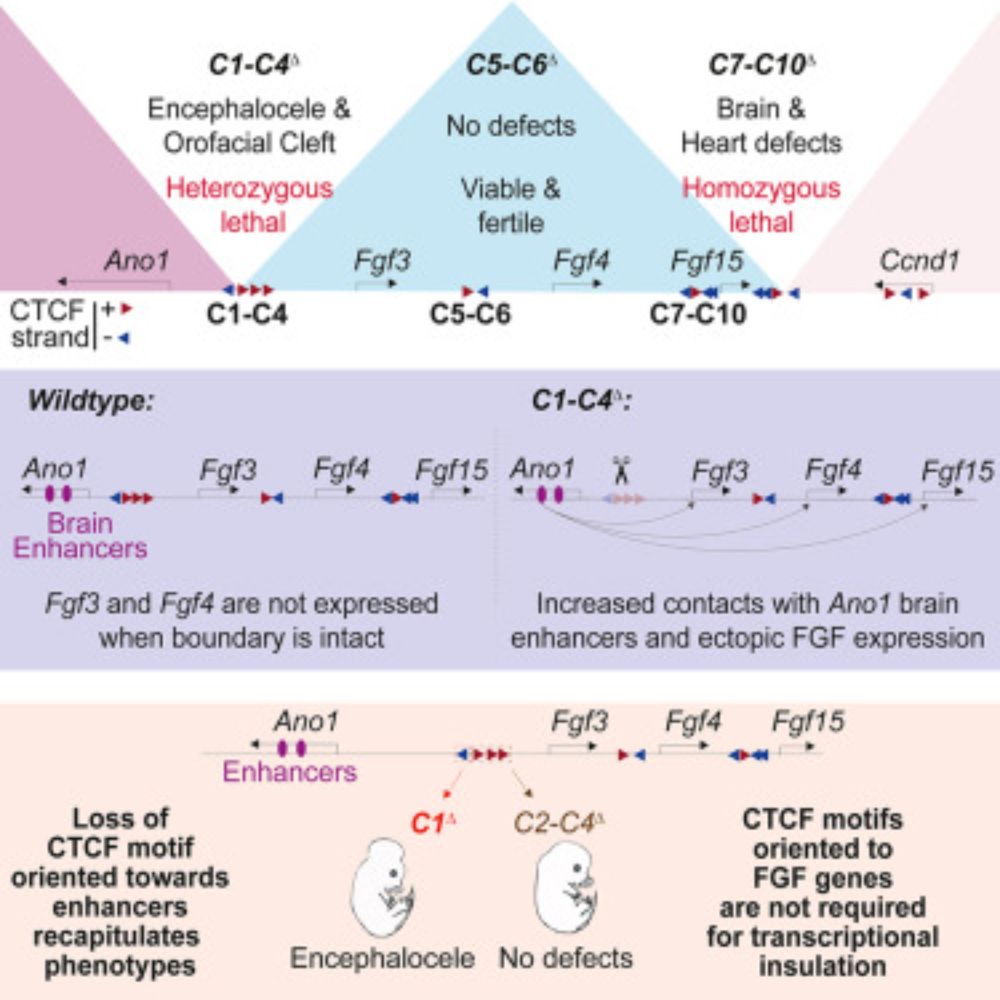
Influenza hemagglutinin subtypes have different sequence constraints despite sharing extremely similar structures
Hemagglutinins (HA) from different influenza A virus subtypes share as little as ∼40% amino acid identity, yet their protein structure and cell entry function are highly conserved. Here we examine the extent that sequence constraints on HA differ across three subtypes. To do this, we first use pseudovirus deep mutational scanning to measure how all amino-acid mutations to an H7 HA affect its cell entry function. We then compare these new measurements to previously described measurements of how all mutations to H3 and H5 HAs affect cell entry function. We find that ∼50% of HA sites display substantially diverged preferences for different amino acids across the HA subtypes. The sites with the most divergent amino-acid preferences tend to be buried and have biochemically distinct wildtype amino acids in the different HA subtypes. We provide an example of how rewiring the interactions among contacting residues has dramatically shifted which amino acids are tolerated at specific sites. Overall, our results show how proteins with the same structure and function can become subject to very different site-specific evolutionary constraints as their sequences diverge. ### Competing Interest Statement JDB consults for Apriori Bio, Invivyd, Pfizer, GSK, and the Vaccine Company. JDB and BD are inventors on Fred Hutch licensed patents related to the deep mutational scanning of viral proteins. National Institute of Allergy and Infectious Diseases, R01AI165821, 75N93021C00015 U.S. National Science Foundation, DGE-2140004 Howard Hughes Medical Institute, https://ror.org/006w34k90
In new work by @jahn0.bsky.social and I in @jbloomlab.bsky.social, we investigate how sequence constraints differ across influenza HA subtypes.
We find ~50% of sites in HA display substantially different amino-acid preferences across H3, H5, and H7.
doi.org/10.64898/202...
21.01.2026 19:22 — 👍 23 🔁 10 💬 1 📌 0
Not that long ago, in vivo mouse enhancer design was a dream. Today, it's a reality! Using transfer deep learning to design de novo synthetic embryonic enhancers active in the heart, limb, and CNS. Great collab with @alex-stark.bsky.social lab! @ucibiosci.bsky.social @impvienna.bsky.social
24.12.2025 15:51 — 👍 76 🔁 23 💬 3 📌 0
Here is a copy of last year's Twitter thread explaining our preprint - jump to (21) for the new stuff 👀
Synergy between cis-regulatory elements can render cohesin dispensable for distal enhancer function
now revised and journal accepted at www.science.org/doi/10.1126/...
🧵👇
27.11.2025 21:58 — 👍 89 🔁 45 💬 4 📌 3
Honored to lead off such an amazing set of speakers 🧬 🌟
02.09.2025 17:47 — 👍 16 🔁 2 💬 1 📌 1
Speaking of T-Rex
02.07.2025 23:21 — 👍 17 🔁 4 💬 0 📌 0
I think the new highest honor in science is an adorable cartoon of your work 🌟
02.07.2025 19:57 — 👍 2 🔁 0 💬 0 📌 0
A dream come true: my first first-author publication! 🧬 🎉 Working to unravel the mysteries of long-range gene regulation has been an incredible journey. Endless gratitude to Evgeny, the lab, and all of the amazing collaborators who helped make this happen 🙌
02.07.2025 19:54 — 👍 70 🔁 14 💬 3 📌 0
How do non-coding variants in enhancers lead to disease? Happy to share our recent work, led by @ewholling.bsky.social, in which we discovered that poised chromatin sensitizes enhancers to aberrant activation by non-coding mutations, contributing to disease. www.biorxiv.org/content/10.1... 1/
23.06.2025 12:56 — 👍 97 🔁 38 💬 3 📌 4

We highlighted new research from the past two years on examples of long-range gene regulation - within large TADs, across TADs, and across chromosomes - comparing mechanisms in flies and mammals 🪰❤️🐁.
www.sciencedirect.com/science/arti...
13.02.2025 12:22 — 👍 29 🔁 8 💬 1 📌 0
It was exciting to dig into all the new discoveries in long range gene regulation while writing this!
22.11.2024 22:27 — 👍 9 🔁 2 💬 0 📌 0
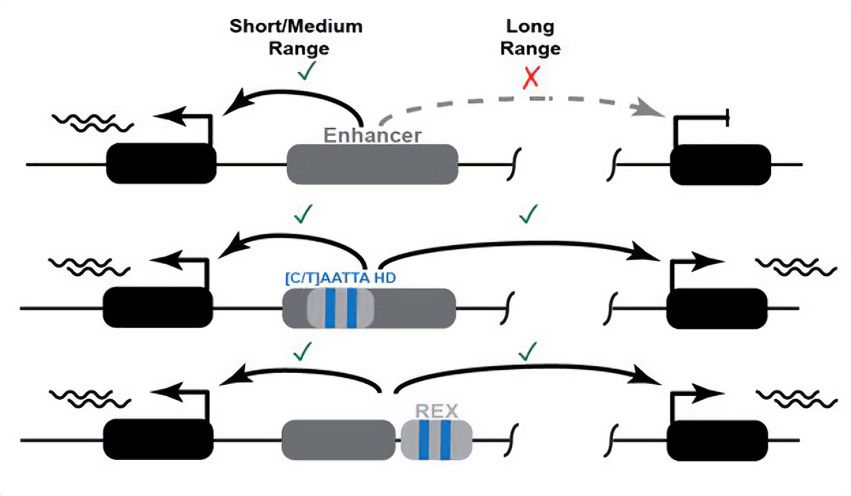
Schematic overview of the proposed mode of action of the newly discovered REX element. Top: An enhancer can activate a gene at a short distance, but not at at long range. Middle: Presence of (C/T)AATTA motifs within an enhancer enable it to act over long distances. Bottom: Coupling a short-range enhancer to the REX element containing the same motifs turns it into a long-range enhancer.
REX - a mammalian "range extender" element that can turn short-distance enhancers into long-distance enhancers.
New preprint from a collaboration led by Grace Bower and Evgeny Kvon.
doi.org/10.1101/2024...
27.05.2024 16:26 — 👍 55 🔁 21 💬 2 📌 4
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
Postdoctoral researcher with @meleckmas.bsky.social at @petermaccc.bsky.social 🇦🇺 | PhD with @vaquerizasjm.bsky.social @mpi-muenster.bsky.social 🇩🇪 | BSc @lcgunam 🇲🇽 | Interested in gene regulation, epigenetics and chromatin architecture.
Fungi 🍄 | Plants 🌵 | Epigenetics 🧬 | Eco-Evo-Devo 🌀 | PhD Student, Stanford 🌲(he/him/il) #FirstGen #QueerinSTEM 🏳️🌈 ♾️
🌎 Science is for everyone.
🍄🟫 Views are my own.
Scientist at IMP in Vienna. Excited about gene expression regulation and its encoding in our genomes - enhancers, transcription factors, co-factors, silencers, AI.
PhD student | Mannervik lab | Stockholm University | developmental epigenetics and chromatin of Drosophila| histone acetylation and methylation
Heliix Brief delivers fast, accessible updates on the latest biomedical research. We scan new papers published in Europe PMC and bring you clear, concise digests focused on relevance and impact.
Molecular biologist. Group leader at MRC LMB Cambridge UK. Fellow at Clare Hall college. #CryoEM #RNAbiology #DNArepair
Views/opinions are my own.
https://www2.mrc-lmb.cam.ac.uk/group-leaders/n-to-s/lori-passmore/
Personal account = views my own. Chromatin/epigenetics.
Science, birds, herps, photography, camera trapping, Warriors, & Cal
Post doc with Mike E Greenberg at HMS, working on molecular neuroscience. Ph.D.at CSHL on identifying a novel family of cell type-specific cofactors. Co-organizer of Gene expression and RNA series, and Fragile nucleosome seminar.
chromatin biology and gene regulation | snRNAseq & scMultiome | Salmonid developmental biology
Core Scientist @roslininstitute @EdinburghUni | @FNucleosome co-organizer
she/her , 🏳️🌈
Skeets can be in English/Turkish/Farsi
🐱🪴📚
https://chromatindynamics.com/
Assistant Professor at Penn State University.
Looking at cool tiny complex things through electromagnetic lens.
Today, chromatin remodeling. Tomorrow? Who knows.
Opinions my own
Assistant prof at OSU|epigenomics of cellular reawakening in yeast & neural stem cells|views mine!|she/her
Diversity, Equity, & Inclusion are values essential to an ethical and modern society|Black lives matter|Trans rights are human rights
PhD in biochemistry and molecular biology. Postdoc at UW-SMPH | Formerly: PhD at Penn State.| Interests: transcription, epigenetics, promoter proximal pausing, cancer, languages, travel. Opinions are my own.
Lab head in stem cell and cancer epigenetics at PeterMac, UniMelb 🇦🇺🧬
Scientist, mum, dreamer
She/her. Views are my own
www.eckmaslab.com
Cancer Biologist.
Big fan of the bone microenvironment.
Chromatin architecture.
Books, coffee, good food and games.
Professor of Neuroscience. Studying neural development, regeneration, and control of innate behaviors at Johns Hopkins.
Assistant professor of Biochemistry and Molecular Biology at Colorado State University. Chromatin in three dimensions. https://swygertlab.colostate.edu/
Assistant Staff Scientist in the Donczew lab at OMRF | broadly interested in how chromatin and other stuff regulate transcription | he/him