Our latest paper has just been published in Cell!
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.
05.11.2025 17:17 — 👍 204 🔁 75 💬 6 📌 10

All but one bZIP monomer, Jun, has significantly higher affinity for other monomers than its own DNA target, favouring the dimer pathway for target search at equilibrium.
Over 10% of human transcription factors are oligomeric. Stoichiometry might be dialled to modulate transcription. We show CREB searches in DNA mixtures as a dimer, and suggest this might be common amongst other members of the bZIP family which fold on binding DNA.
www.biorxiv.org/content/10.1...
08.09.2025 09:36 — 👍 21 🔁 3 💬 1 📌 0

Activity of most genes is controlled by multiple enhancers, but is there activation coordinated? We leveraged Nanopore to identify a specific set of elements that are simultaneously accessible on the same DNA molecules and are coordinated in their activation. www.biorxiv.org/content/10.1...
18.08.2025 12:23 — 👍 95 🔁 39 💬 2 📌 2

Disruption of the SAGA CORE triggers collateral degradation of KAT2A
The SAGA transcriptional co-activator complex regulates gene expression through histone acetylation at promoters, mediated by its histone acetyl transferase, KAT2A. While its structure and function ha...
I am pleased to share a new preprint from my postdoc in the @dseruggia.bsky.social lab. We discovered a structural dependency within the CORE module of the transcriptional coactivator SAGA, which regulates the stability of its histone acetyl transferase, KAT2A.
www.biorxiv.org/content/10.1... 1/n
01.08.2025 17:42 — 👍 10 🔁 3 💬 1 📌 1
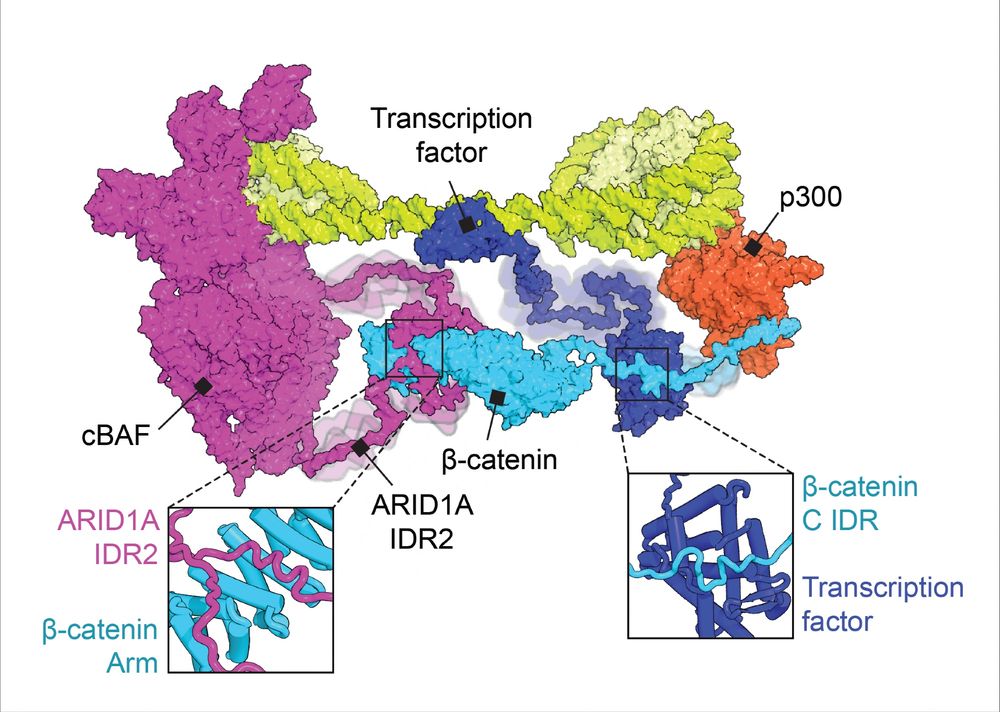
How do chromatin remodelers use #IDRs to find TF binding partners? In our new Molecular Cell paper, we show that β-catenin is an adaptor that links SWI/SNF (cBAF) subunit ARID1A with binding partners via IDR-domain interactions.
www.cell.com/molecular-ce...
22.07.2025 18:42 — 👍 89 🔁 34 💬 11 📌 4

Celebrating 10 years of our lab with a new preprint:
www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth
29.03.2025 12:45 — 👍 149 🔁 58 💬 6 📌 2

Gallo et al. describe NF-YAr, a retroposed copy of the NF-YA transcription factor subunit, in several mammal lineages. Likely expressed in sperm, NF-YAr shows structural changes that suggest altered regulatory function.
🔗 doi.org/10.1093/gbe/evaf071
#genome #evolution #transcription #TEsky
27.05.2025 08:09 — 👍 4 🔁 2 💬 0 📌 0
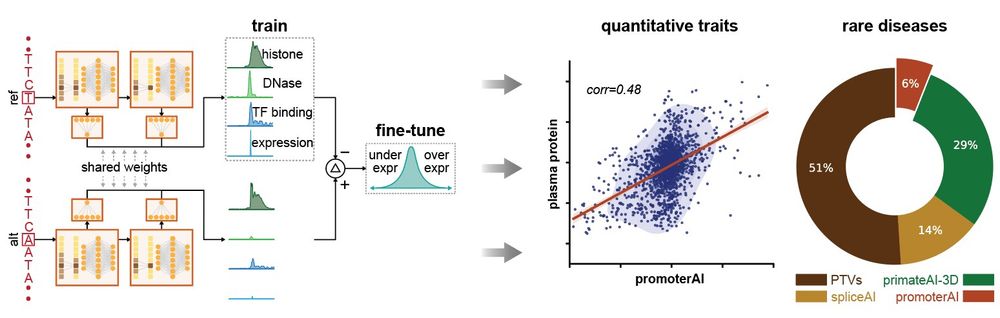
Excited to share my first contribution here at Illumina! We developed PromoterAI, a deep neural network that accurately identifies non-coding promoter variants that disrupt gene expression.🧵 (1/)
29.05.2025 23:57 — 👍 60 🔁 21 💬 1 📌 1
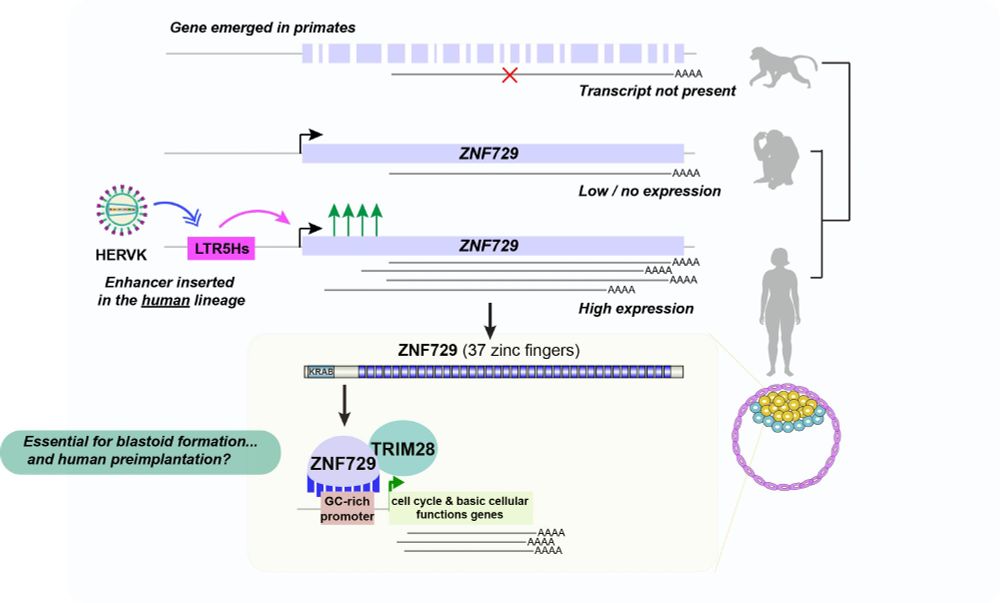
I am thrilled to share our preprint where we take advantage of the unique opportunities offered by human blastoids to uncover a human-specific mechanism potentially playing a role in preimplantation 🧵. www.biorxiv.org/content/10.1...
12.05.2025 20:24 — 👍 73 🔁 32 💬 3 📌 2

Design principles of cell-state-specific enhancers in hematopoiesis
Screen of minimalistic enhancers in blood progenitor cells demonstrates widespread
dual activator-repressor function of transcription factors (TFs) and enables the model-guided
design of cell-state-sp...
Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
08.05.2025 16:06 — 👍 139 🔁 58 💬 4 📌 8
Congratz Tanja!!
27.04.2025 20:30 — 👍 1 🔁 0 💬 1 📌 0

Large language models trained on DNA sequences, also known as genomic language models (gLMs), hold significant potential to advance our understanding of genomes and the interactions between DNA elements that drive complex functions. In this issue of Trends in Genetics, Benegas et al. review key opportunities and challenges for gLMs, outlining important considerations for their development and evaluation to benefit the genomics community. In this image, the two binary strings correspond to reverse-complementary DNA sequences (00 = A, 01 = C, 10 = G, and 11 = T). The connecting rectangles represent “embeddings” learned by gLMs. Illustration by Yun S. Song.
TiG’s April issue is live!
Topics include genomic language models, hyperdivergent regions, meiotic DSBs, cfDNA in pop’n genetics, genital evolution, ‘rulers’ for TSS selection, spatial multiomics in embryos, and more:
www.cell.com/trends/genet...
Thanks to @yun-s-song.bsky.social for the cover!
08.04.2025 13:30 — 👍 7 🔁 1 💬 1 📌 0
Having a stressful day? Lay back, crack a beer open and read about how chromatin accessibility responds to perturbations of TF binding at the single molecule level bit.ly/3XQMFxN. I'm incredibly thankful to my PhD mentors @arnaudkr.bsky.social and Judith Zaugg for these super productive years!!
08.04.2025 15:41 — 👍 15 🔁 6 💬 2 📌 0
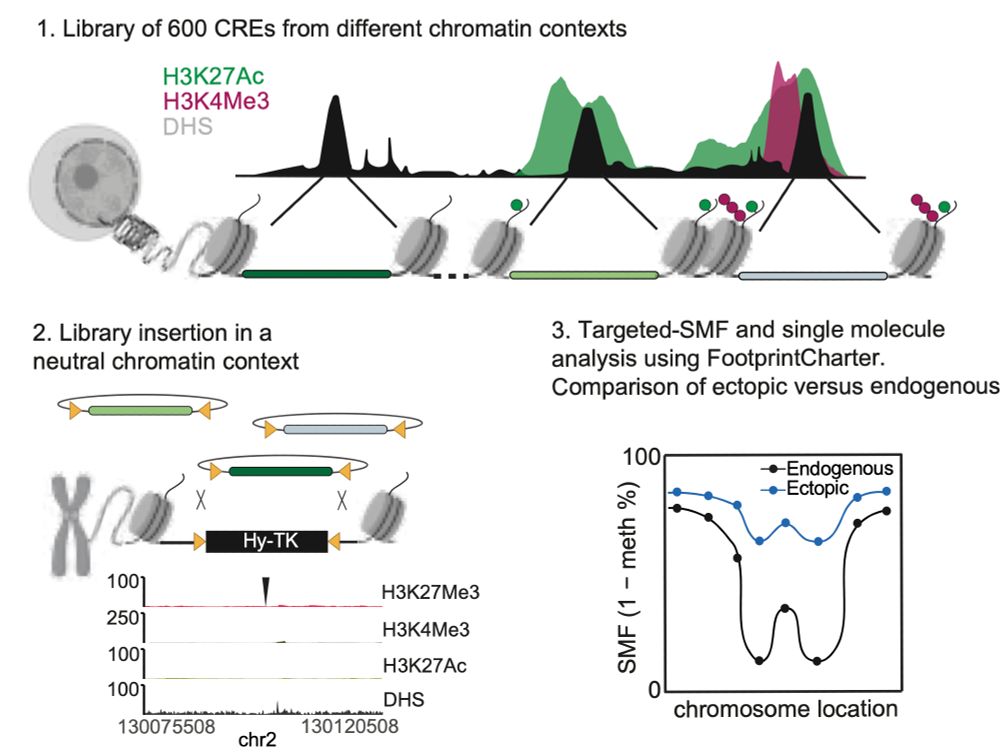
Happy to share the latest story from @arnaudkr.bsky.social's lab @embl.org! With @guidobarzaghi.bsky.social, we used Single Molecule Footprinting to quantify how often chromatin is accessible at enhancers after TF and chromatin environment changes! Check our preprint bit.ly/3XQMFxN + thread ⬇️ 1/11
08.04.2025 13:51 — 👍 78 🔁 33 💬 4 📌 2
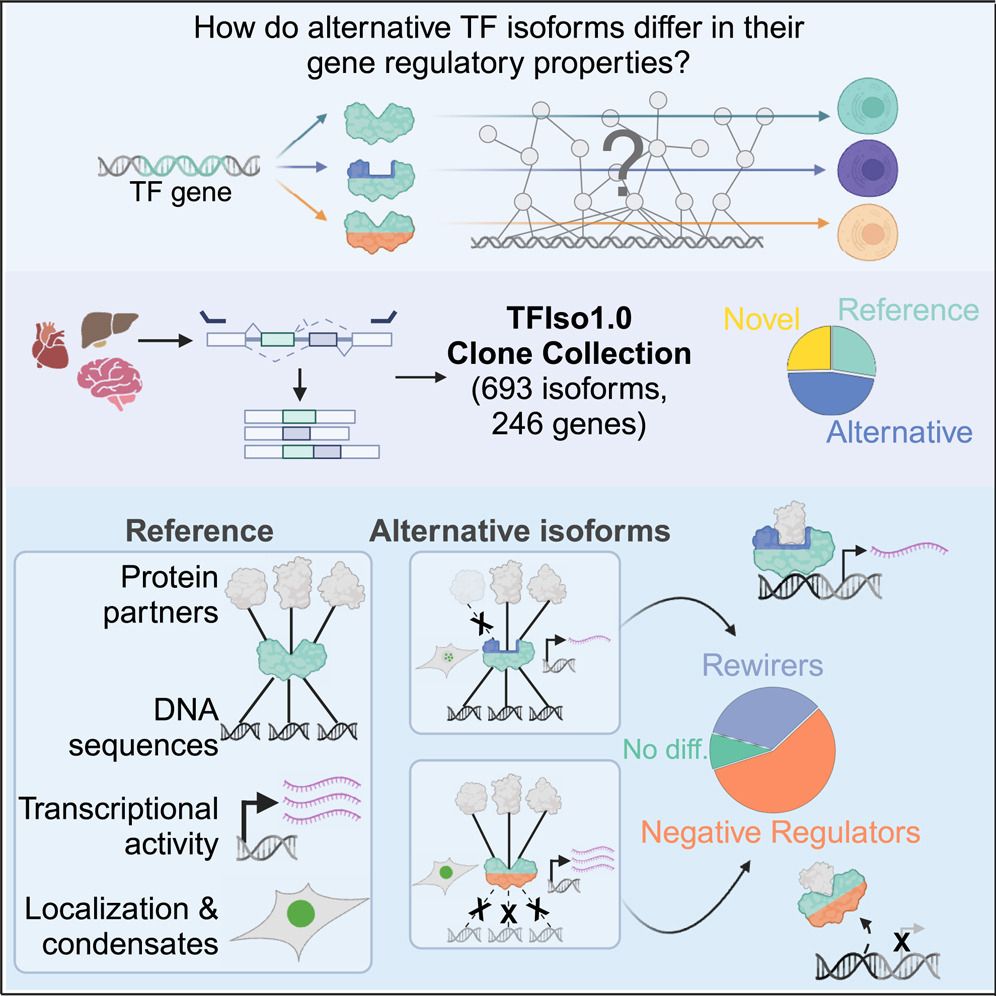
our work on the molecular differences between transcription factor isoforms is out now in Molecular Cell!
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
26.03.2025 17:10 — 👍 159 🔁 70 💬 7 📌 5
Read our collaborative paper
@longchrom.bsky.social and @molinalab.bsky.social
showing that disruption of NuA4 and ATAC coactivator complexes cause gene-specific transcript buffering, suggesting that cells dynamically adjust RNA export/degradation to sustain cellular homeostasis.
21.03.2025 17:27 — 👍 11 🔁 6 💬 3 📌 0
What is the multiscale structure of chromatin condensates? How does it shape thermodynamic and material properties?
We address this at near-atomistic resolution🔥🔥🔥 using cryoET (Rosen & Villa labs, led by H Zhou), a new multiscale model (K Russell) and cryoET-guided sims (J Huertas & J Maristany)
23.01.2025 09:32 — 👍 117 🔁 30 💬 7 📌 4

Multiscale footprints reveal the organization of cis-regulatory elements www.nature.com/articles/s41...
▶️Footprints of DNA–protein interactions from bulk & single-cell ATAC-seq ⤵️
▶️Deep learning to infer transcription factor and nucleosome binding
▶️Age-associated reduction of nucleosome footprints
23.01.2025 00:49 — 👍 16 🔁 7 💬 0 📌 0
The EU channel for scientists & researchers: opportunities to fund your ideas, updates from EU-funded science, and scientific resources you can use in your work.
Our long-term research goal is to understand and predict gene regulation based on DNA sequence information and genome-wide experimental data.
Biophysicist who loves disordered proteins and transcription. PI at Univ of Oxford, UK. She/her.
MBE publishes fresh insights into the patterns and processes that impact the evolution of life at molecular levels.
🔗 academic.oup.com/mbe
🏠 @official-smbe.bsky.social
🤝 @genomebiolevol.bsky.social
#evobio #molbio #science #biology #societyjournal
Love predicting genomic things. Postdoc @crgenomica.bsky.social at the Probabilistic Machine Learning and Genomics group.
Creator of @splicingnews.bsky.social
Structural and Molecular biologist | Postdoc at MPI-NAT @mpi-nat.bsky.social Patrick Cramer‘s lab @patrick-cramer.maxplanck.de | Cryo-EM | DNA-protein interactions
Baylor College of Medicine
PhD student in the Brummelkamp lab at the Netherlands Cancer Institute.
Passionate about science and sarcasm. 🏳️🌈
GBE publishes leading original research at the interface between evolutionary biology and genomics.
🔗 academic.oup.com/gbe
🏠 @official-smbe.bsky.social
🤝 @molbioevol.bsky.social
#genome #evolution #science #biology #societyjournal
(she/her) Computational biologist and post-doc scientist in the Greenleaf and Kundaje labs at Stanford. Interested in understanding how cells know what to become (transcription factors, gene regulation, dev bio, open science) www.selinjessa.com
NYU postdoc with Tsirigos and Aifantis labs.
Former EMBL Heidelberg PhD with Krebs and Zaugg labs.
PhD student @EMBL in the Krebs Lab
K99/R00 postdoc, 24/7 feminist @ Harvard/BWH. generally fascinated by genomes. big fan of reality tv, good writing, crosswords, and cats. obsessed with my e-bike. she/her
Banerjee lab postdoc. Molecular biologist. Interested in biomolecular condensates and regulation of gene expression.
Physics at Ulm University | live-cell/organism single-molecule microscopy | super-resolution microscopy | gene regulation | chromatin architecture | embryo development
A reviews journal from Cell Press that fosters an appreciation for advances being made on all fronts of genetic research.
Editor: Maria Smit
https://www.cell.com/trends/genetics/home
François Wallace Monahan Fellow, MSKCC (NYC). Previously Johns Hopkins 🇺🇸 and IGBMC 🇫🇷. Views my own.
Scientist interested in genes in living cells. Associate Professor at Colorado State University.
Postdoc at @CBD KU Leuven 🇧🇪. Interested in translation, structural biology, ageing and everything involving proteins
Theoretical and computational biophysics | Postdoc at Hummer's lab
@hummerlab.bsky.social | PhD at @mpibp.bsky.social | MSc. @unistuttgart.bsky.social | Physics at @uniandes.bsky.social 🇨🇴 🇩🇪 🇪🇺