Scientists at the NKI have made a remarkable discovery: a new protein duo that can turn genes on and off. They have named this duo Zincore. Together, these proteins help other proteins bind more firmly to the DNA, allowing them to perform their functions more effectively www.nki.nl/news-events/...
10.07.2025 13:39 — 👍 12 🔁 1 💬 1 📌 0
🚀 My first Cryo-EM structures are OUT on the PDB!
🎉Super excited about this milestone as PhD, it’s been a journey, and I’m grateful to finally have these as my first Cryo-EM structures! 🙌
🔁Thank you for sharing!❤️
Read @daaninthelab.bsky.social et. al.: www.science.org/doi/10.1126/...
⬇️ 2nd video
09.07.2025 22:43 — 👍 55 🔁 13 💬 2 📌 0
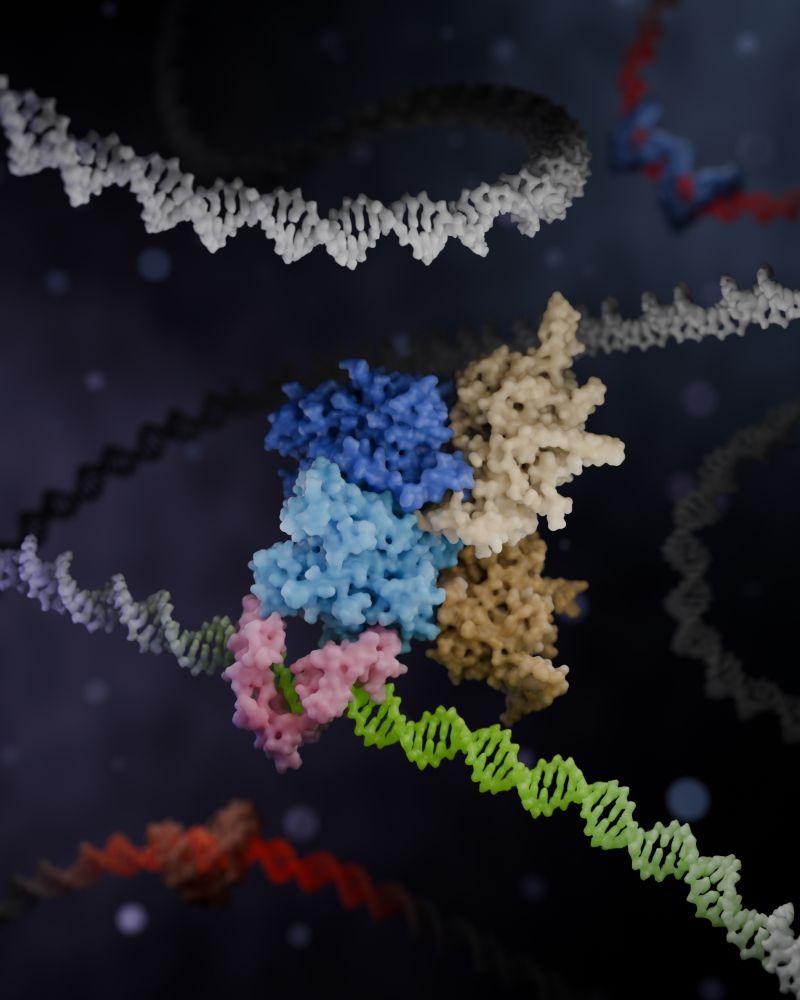
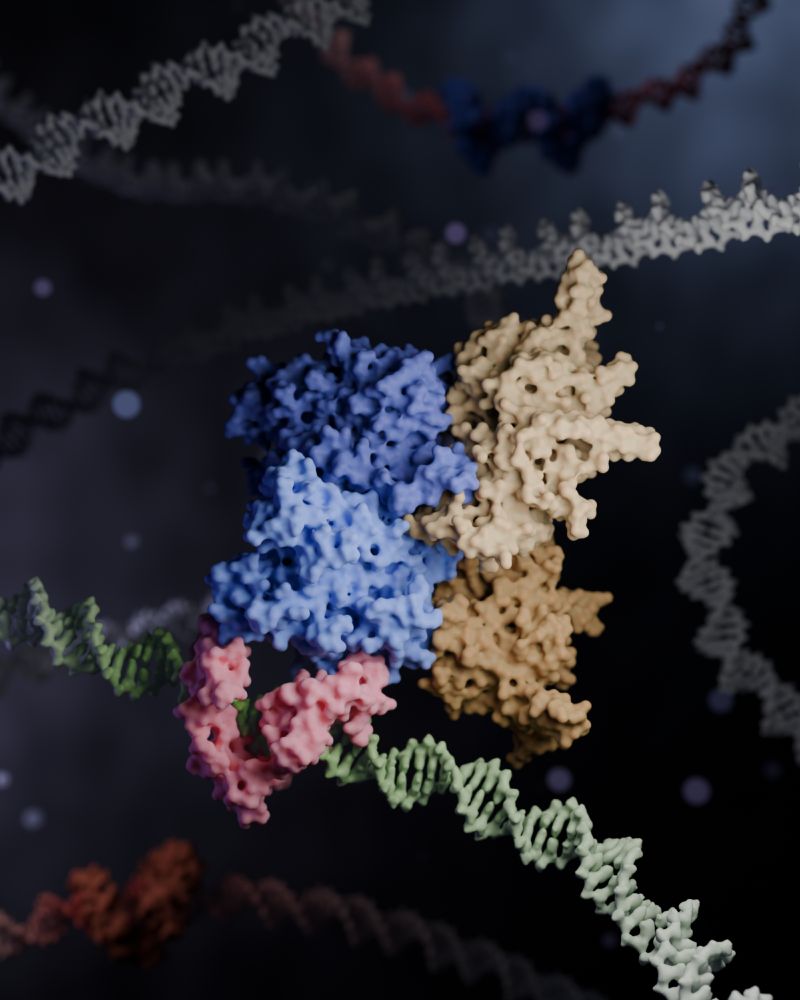
3D render of a Zincore complex locking ZFP91 onto DNA, promoting gene activation.
Made in Blender with Molecular Nodes.
📢 Ready for this? A new way to think about Gene Regulation!
🚨A new coregulator complex, Zincore, acts as a "MOLECULAR GRIP", stabilizing TFs at DNA binding sites across the genome!🧬
💪Proud to be part of this work led by @daaninthelab.bsky.social
Read more: science.org/doi/10.1126/science.adv2861
08.07.2025 11:18 — 👍 34 🔁 12 💬 1 📌 0
Thanks a lot Filip, the perspective was really a wonderful surprise!
05.07.2025 14:05 — 👍 1 🔁 0 💬 0 📌 0
oops: a novel protein complex*
04.07.2025 14:39 — 👍 0 🔁 0 💬 0 📌 0
& blueskyless: Erica De Zan, Guizela Huelsz-Prince, Marleen Dekker, Alex Fish, Lona Kroese, Simon Linder, Miguel Hermandez-Quiles, Ji-Ying Song, Lodewyk Wessels, Sebastian Nijman 11/11
04.07.2025 11:08 — 👍 3 🔁 0 💬 0 📌 0
Thanks to all coauthors: @rborza.bsky.social @sgregoricchio.bsky.social @amazouzi.bsky.social @zwartlab.bsky.social @lindersimon.bsky.social @tassosperrakis.bsky.social @thijnbrummel.bsky.social
04.07.2025 11:07 — 👍 7 🔁 0 💬 1 📌 0
Please check the paper for more experiments and interesting findings and stay tuned, as I am convinced this fascinating complex will be connected to much more interesting biology in the future. 10/11
04.07.2025 11:05 — 👍 2 🔁 0 💬 1 📌 0
I'm truly excited to share this work with you today. Unraveling this fascinating complex was a rewarding challenge, made possible by the incredible teamwork behind it. I'm deeply grateful to all the brilliant collaborators who contributed their insights and energy to make this possible. 9/11
04.07.2025 11:05 — 👍 3 🔁 0 💬 1 📌 0
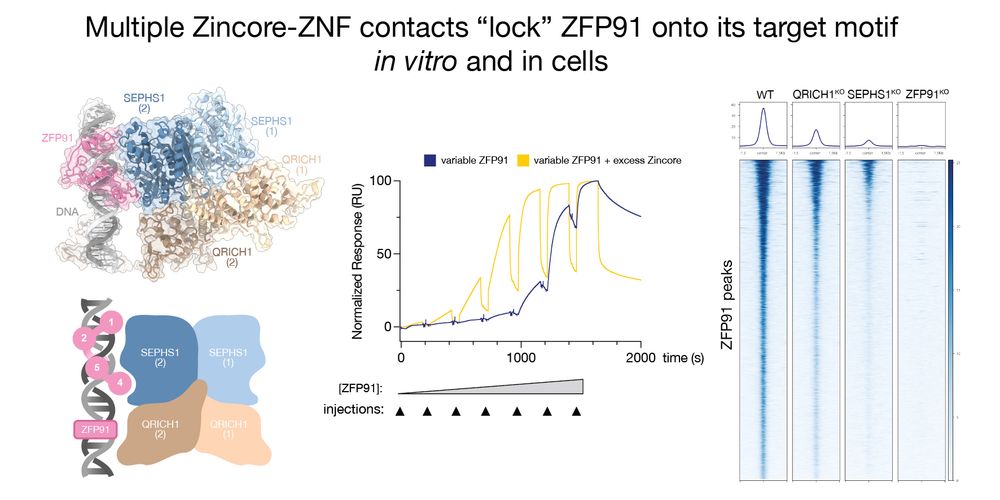
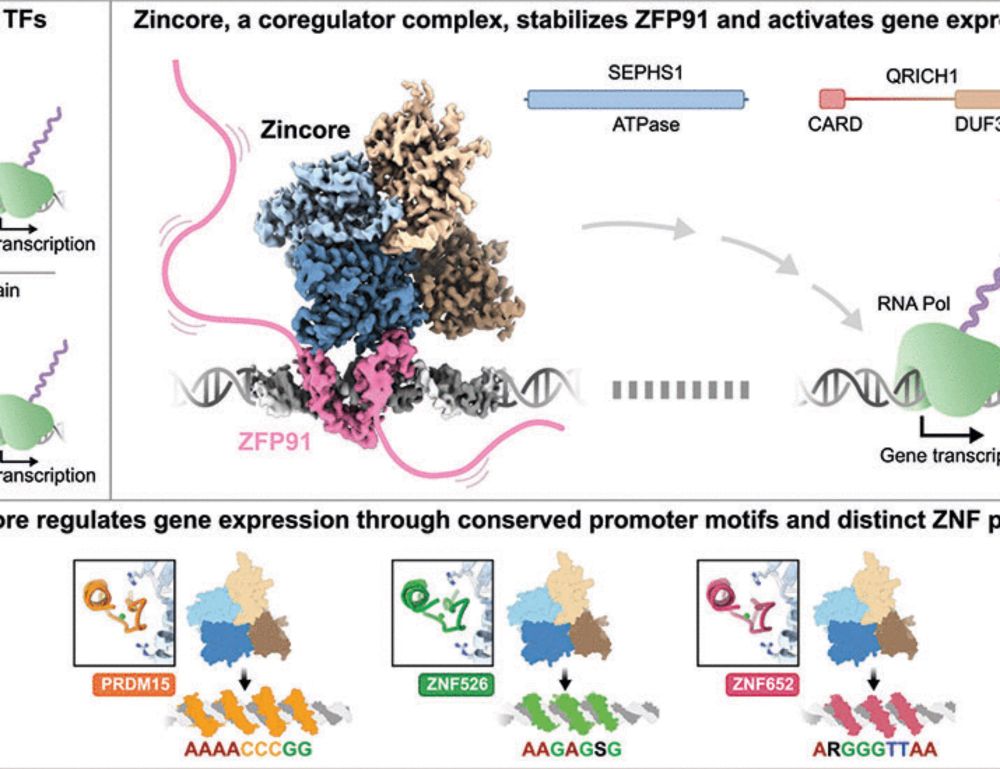
1) Zincore recognises the highly conserved ZNF fold, explaining its broad ZNF recognition, and 2) Zincore locks the ZNF onto DNA, stabilizing promoter binding. Revealing an entirely new mode of how ZNFs activate transcription. 8/11
04.07.2025 11:05 — 👍 3 🔁 0 💬 1 📌 0
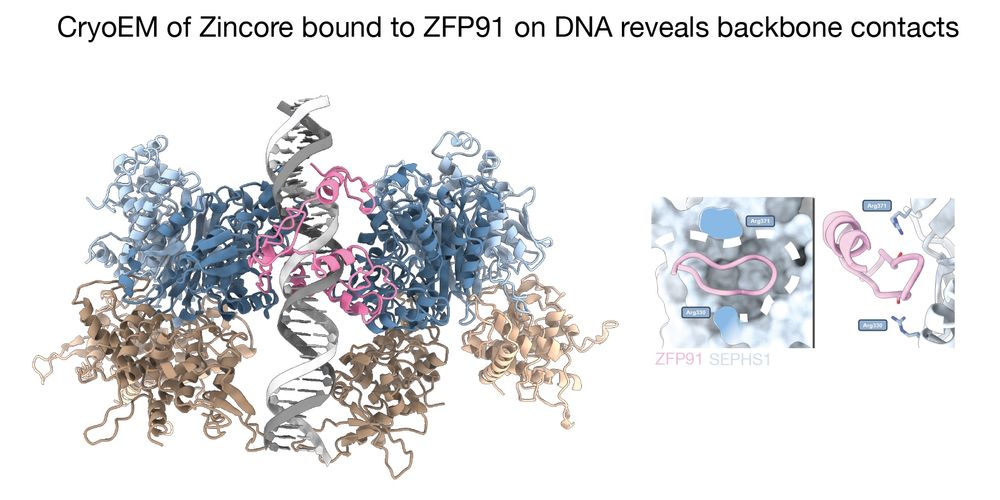
CryoEM analysis of the Zincore bound to ZFP91 onto the DNA revealed a striking complex: instead of recognising an effector domain, Zincore binds to multiple DNA-bound ZNF domains directly, which revealed two peculiarities: 7/11 @rborza.bsky.social @tassosperrakis.bsky.social
04.07.2025 11:04 — 👍 6 🔁 0 💬 1 📌 1
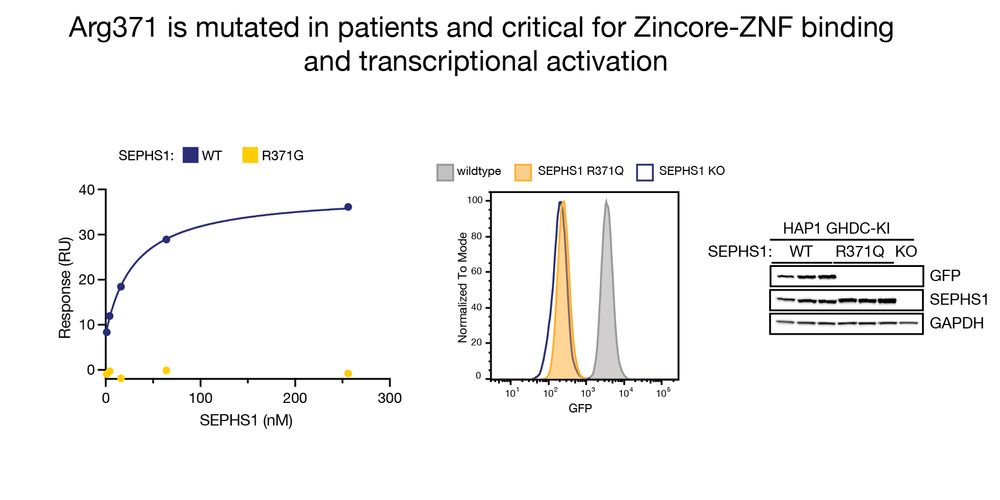
Additionally, in vitro binding experiments demonstrated direct binding of Zincore to ZNFs on DNA. This depended on SEPHS1 Arg371, a residue which is mutated in patients with a neurodevelopment syndrome, and turned out to be critical for transcriptional activation. 6/11
04.07.2025 11:03 — 👍 4 🔁 0 💬 1 📌 0
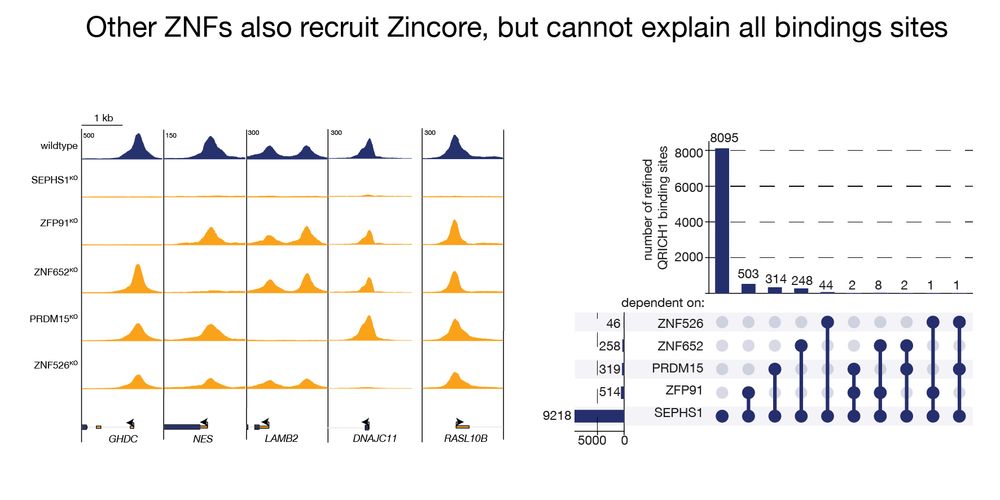
Additional searches revealed different transcription factors to bring Zincore to chromatin: coincidentally all zinc finger transcription factors? 5/11
04.07.2025 11:02 — 👍 4 🔁 0 💬 1 📌 0
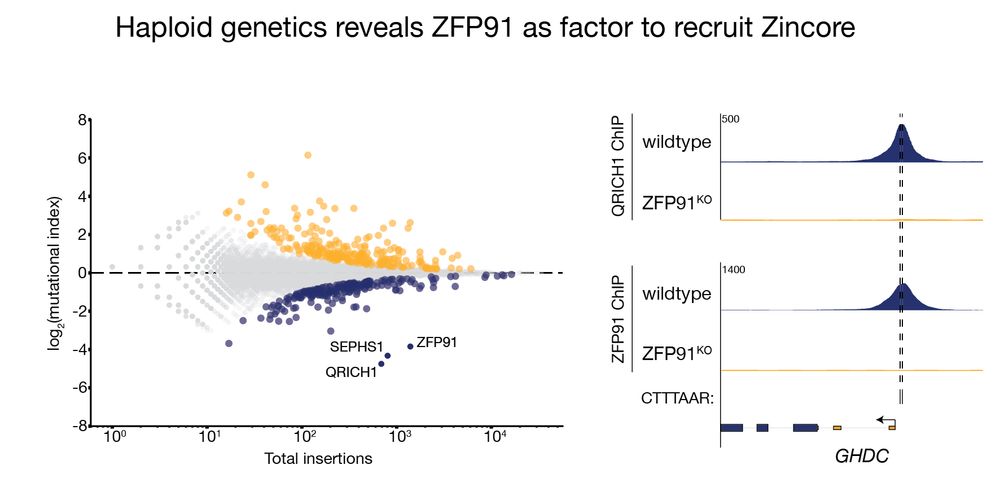
Using haploid genetics in this reporter cell line identified ZFP91, a zinc finger transcription factor, as Zincore partner. ChIP-seq showed Zincore is directly involved in gene expression via recruitment to chromatin by ZFP91, though this explains only a fraction of Zincore promoter binding. 4/11
04.07.2025 11:01 — 👍 5 🔁 0 💬 1 📌 0
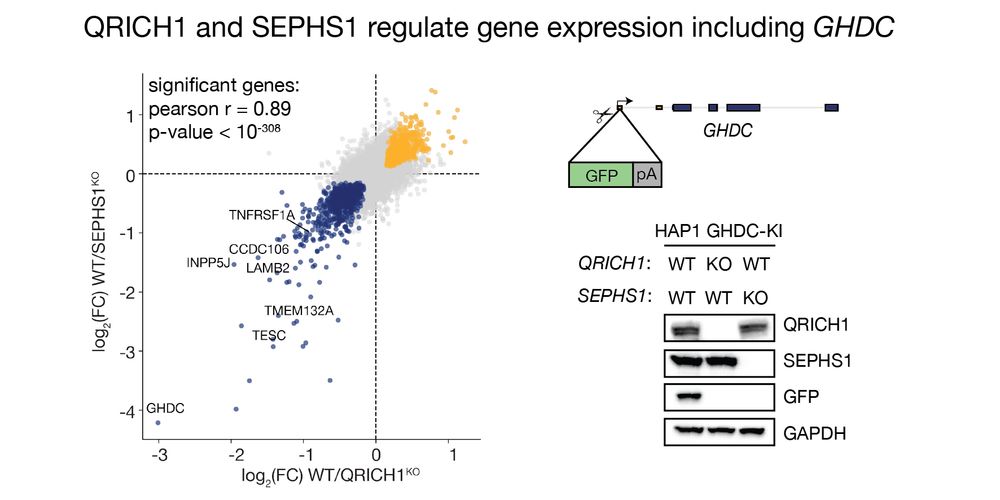
As a PhD student, this was challenging. I worked on an uncharacterized protein complex, yet had no idea how it worked. RNAseq showed many genes were activated by QRICH1 and SEPHS1, but how? This gave us options! We made a reporter in one of Zincore's activated genes: GHDC. 3/11
04.07.2025 11:01 — 👍 3 🔁 0 💬 1 📌 0
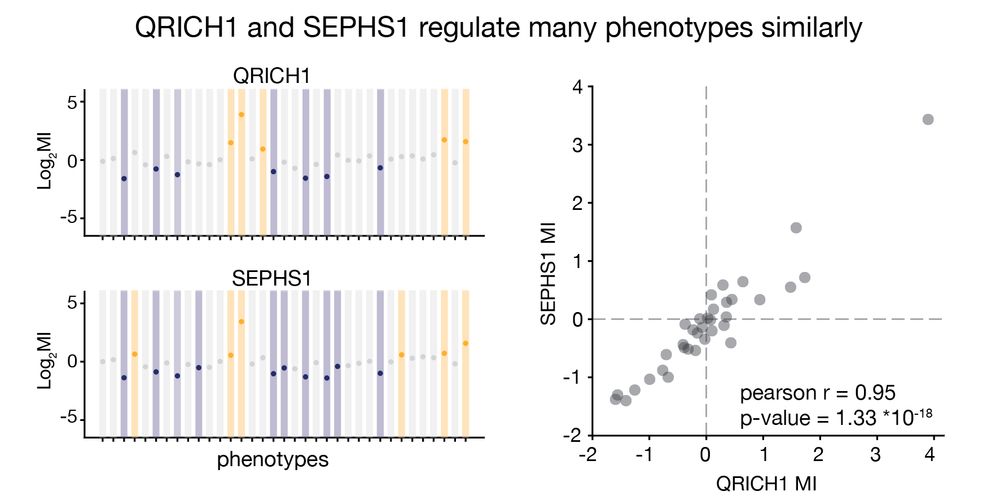
From analysing haploid genetic screens carried out by our group for different phenotypes we found that QRICH1 and SEPHS1 affect phenotypes similarly. They form a complex that we name Zincore. 2/11
04.07.2025 11:00 — 👍 4 🔁 0 💬 1 📌 0
Assistant Professor of Cell Biology at Harvard Medical School | HHMI Freeman Hrabowski Scholar | Nexus of chromatin, transcription, replication, and epigenetics. farnunglab.com
Assistant Prof at Utrecht University, studying the process of transcription elongation vs early termination. (she/her)
Group Leader and Lecturer at the Australian National University | researching bacterial thiol redox pathways and virulence factors | community builder | storyteller | views my own | she/her
Associate professor and CRUK Senior Fellow, University of Copenhagen and University of Oxford. My lab researches how DNA repair defects and loss of genome stability cause disease, including cancer.
https://icmm.ku.dk/english/research-groups/blackford-group
Senior editor @science.org. Molecular biology, #DNA, #RNA, #gene regulation, #epigenetics, nuclear biology, #chromatin biology, 3D #genome, #synbio, #CRISPR and gene editing, other bacterial immune systems, and #AI in all these
PhD student in the van Steensel and Medema labs | Netherlands Cancer Institute
Postdoc in Bas van Steensel Lab, Netherlands Cancer Institute | previously PhD with Luca Giorgetti, Friedrich Miescher Institute
Genome Architecture | Gene regulation | Live-cell imaging | Genomics
Postdoc at TU Delft (Cees Dekker lab) || PhD at NKI, Amsterdam (Benjamin Rowland lab) || Chromosome biology enthusiast, fascinated by SMC complexes 🧬➰🍩
Group leader at MRC Human Genetics Unit, University of Edinburgh. Interdisciplinary research on disease #epigenetics. Part-time solo dad. Occasional music and climbing.
https://institute-genetics-cancer.ed.ac.uk/research/research-groups-a-z/sproul-group
Group Leader - Reader (Assoc. Professor) in AI and Cancer Epigenetics at Blizard Institute, Queen Mary University of London.
#GeneRegulation, #Chromatin, #Epigenetics, #AI, #bioinformatics
https://www.qmul.ac.uk/blizard/all-staff/profiles/radu-zabet.html
Chair, Computational BIology and Medicine Program, Princess Margaret Cancer Centre, University Health Network.
Associate Professor, Medical Biophysics, University of Toronto.
Disclosures: https://github.com/michaelmhoffman/disclosure/
Chromatin and cryoEM afficionada. Still a fan of crystallography. Avid Colorado hiker. Will call out ugly nucleosome cartoons. Come for the science, stay for the mountain pictures and snark. Opinions and snark are my own.
Professor at KTH, NY Genome Center, SciLifeLab, working on functional genomics and human genetics.
Resident panhandler, http://trichelab.org/
Posts may contain trace quantities of blood🩸, chromatin 🧬, and stats 🧮
CV: https://scholar.google.com/citations?user=AOoIO74AAAAJ
Views expressed are my own (but for the right price they can be yours!)
Molecular evolution, chromatin, archaea and oddball biology. Associate Professor @oxfordbiochemistry.bsky.social Fellow @trinityoxford.bsky.social
Reproductive biologist 🇵🇹 in 🇩🇪
Institute of Reproductive Genetics, CMG, Uni-Münster
PI CRU326 "Male Germ Cells"
DNA methylation, spermatogenesis, ageing, male infertility
Hobby collector, slowest runner in the west
https://linktr.ee/SandraLaurentino
Ahh well-a everybody's heard
about the NuRD.
#NuRDistheWord
Cycling/Chromatin Remodelling/Transcription/Enhancers/StemCells
We've got it all.
Living Systems Institute, University of Exeter
https://lsi.exeter.ac.uk/groups/hendrich-group/
RNAPII &Transcription Regulation -Chromatin Conformation & Nuclear Architecture - ESCs & Neuronal Differentiation - Nuclear Mechanotransduction - Cell Plasticity
UMG, Göttingen, Germany