
Best practices and tools in R and Python for statistical processing and visualization of lipidomics and metabolomics data www.nature.com/articles/s4...
01.10.2025 14:15 — 👍 10 🔁 3 💬 0 📌 0@mheming.bsky.social
mheming.com

Best practices and tools in R and Python for statistical processing and visualization of lipidomics and metabolomics data www.nature.com/articles/s4...
01.10.2025 14:15 — 👍 10 🔁 3 💬 0 📌 0
Neuron-reactive KIR+CD8+ T cells display an encephalitogenic transcriptional program in autoimmune encephalitis @natcomms.nature.com
www.nature.com/articles/s41...
Do you use agent mode? Because then copilot often corrects itself. I often write some code using the package I want, e.g. Seurat. And then prompt e.g. Write functions to read in all other samples, don t repeat yourself. That works quite well. Sometimes needs a second prompt.
23.09.2025 04:59 — 👍 1 🔁 0 💬 0 📌 0
A population of anti-CD20–resistant dural B cells that remain in the mouse brain parenchyma at disease remission in progressive multiple sclerosis @jengommerman.bsky.social @jem.org
rupress.org/jem/article-...
2024 MS McDonald criteria now published
www.thelancet.com/journals/lan...

Really insightful post from Julie Tibshirani (spotted in LinkedIn, can't find on Bsky) reflecting on #rstats 's unique governance structure and what can be learned for other languages
jtibs.substack.com/p/if-all-the...

I was loving Claude Code... until I tried it with #rstats. Constant errors, wouldn't use the tidyverse even when asked, "optimized" functions were slower.
Frustrated, I started a session just to teach R to Claude and summarize what it learned into a CLAUDE.md file gist.github.com/sj-io/3828d6...

A thumbnail with a dark background and the VS Code logo that reads "What's new in Visual Studio Code August Update 1.104. Add context from AGENTS.md, confirm agent edits for sensitive files, improved chat UX experience, control chat modes from prompt files, improved terminal command approvals, automatic model selection, model flexibility via VS Code extensions, preview and merge Git worktree changes, configurable inline suggestions delay, and collaborate with background coding agents in Chat Sessions view.
🚀 v1.104 of VS Code is here! Check out what's new:
🤖 Improved coding agent integration
📄 AGENTS.md file support for better context
🔍 New Auto mode (Preview) for smart model selection
🔑 Model flexibility via BYOK extension API
…and more: aka.ms/VSCodeRelease
Here are the highlights 🧵

I am beyond excited to announce that ggplot2 4.0.0 has just landed on CRAN.
It's not every day we have a new major #ggplot2 release but it is a fitting 18 year birthday present for the package.
Get an overview of the release in this blog post and be on the lookout for more in-depth posts #rstats
23 tools to work with (single-cell) TCR/BCR-seq immune repertoire data 🧵 👇
08.09.2025 14:15 — 👍 10 🔁 4 💬 1 📌 0You should include quarto, which also includes python and julia and includes bookdown, blog down etc per se
07.09.2025 16:22 — 👍 1 🔁 0 💬 0 📌 0Very cool. Habe you tried Kinesis Advantage before for a direct comparison? Voyager looks stylisher, although Kinesis has this "bowl" area and more keys in the thumb cluster (which i like for Alt, Ctrl, Super, Alt-Gr, Space, Backspace) .. voyager looks better for a voyage :)
01.09.2025 19:14 — 👍 0 🔁 0 💬 1 📌 0Nice. Which keyboard is that? I am quite happy with kinesis advantage but that one looks also comfortable.
01.09.2025 17:22 — 👍 0 🔁 0 💬 1 📌 0BOOM! Cool to see the final form of this great work now published. Deep new insight into human peripheral neuropathy. Congrats @mheming.bsky.social and colleagues on a great study.
24.08.2025 15:21 — 👍 11 🔁 5 💬 0 📌 0Very impressed with this finding which is quite similar to what we have also seen in DPN nerves. Suggests a completely different disease modifying approach for multiple types of peripheral neuropathy.
24.08.2025 15:23 — 👍 3 🔁 1 💬 0 📌 0Linking people who were involved or who might be interested in reading this:
@mzhlab.bsky.social @clsommer25.bsky.social
@tommytang.bsky.social @tedpricethepainguy.bsky.social @denklab.bsky.social

13/13 Validated in a larger cohort (370 fascicles; 18 CTRL, 12 CIDP): manual perineurium measurements. Linear mixed model (age+sex fixed, center/sample random) showed perineurial area & perimeter increased in CIDP.
24.08.2025 13:07 — 👍 1 🔁 0 💬 1 📌 0
12/13
PNPs showed previously unknown perineurial hyperplasia and fibrotic dispersion and this was most pronounced in immune-mediated PNPs.

11/13 Transcriptional changes affected multiple cells outside of the endoneurium across PNPs, suggesting PNPs as pan-nerve diseases (using miloDE)
24.08.2025 13:07 — 👍 0 🔁 0 💬 1 📌 0
10/13 immune, repairSC and periC3 cluster also correlated with clinical disease severity (INCAT score) and myelin thickness (g-ratio). (using CNA @ilyakorsunsky.bsky.social
24.08.2025 13:07 — 👍 0 🔁 0 💬 1 📌 0
9/13 LAM markers (SPP1/osteopontin, PLIN2, CD36, FABP5) colocalized histologically with myelin and BODIPY (labels neutral lipids). LAM thus likely has myelin-phagocytizing capacity.
24.08.2025 13:07 — 👍 1 🔁 0 💬 1 📌 0
8/13 PNPs shared an increase in endoneurial lipid-associated macrophages (LAM).
24.08.2025 13:07 — 👍 0 🔁 0 💬 1 📌 0
7/13 Human peripheral nerves contained complex immune cells, including a plethora of myeloid cell populations. Macrophage clusters exhibited an endoneurial to epineurial gradient, previously described in rodents.
24.08.2025 13:07 — 👍 0 🔁 0 💬 1 📌 0
6/13 Integrating spatial transcriptomics with snRNA-seq provided an unbiased histological annotation of peripheral nerve cells.
24.08.2025 13:07 — 👍 0 🔁 0 💬 1 📌 0
5/13 We identified novel and human-specific nerve cell type markers using snRNA-seq, which were validated using spatial transcriptomics
24.08.2025 13:07 — 👍 0 🔁 0 💬 1 📌 0
4/13 Single nuclei sequencing dissected cell populations in the sural nerve in great detail, replicating and extending cell populations identified in single cell rodent studies. Interestingly, we identified transcriptionally heterogeneous perineurial fibroblasts.
24.08.2025 13:07 — 👍 0 🔁 0 💬 1 📌 03/13 We combined single nuclei sequencing of 33 human polyneuropathy patients and four controls (365,708 nuclei) with subcellular spatial transcriptomics (#Xenium).
24.08.2025 13:07 — 👍 0 🔁 0 💬 1 📌 02/13 Polyneuropathies (PNP) are among the most common neurological diseases (~4%, but the underlying causes remain unclear in many patients (~20-30%). We used state of the art single cell technologies to better characterize sural nerves in health and disease.
24.08.2025 13:07 — 👍 0 🔁 0 💬 1 📌 0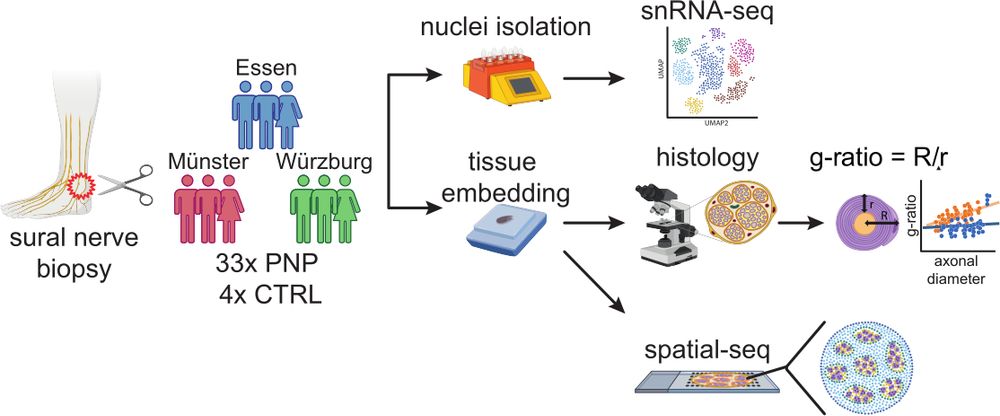
Very proud that our human PNS atlas is finally published in @natcomms.nature.com We found perineurial hyperplasia and lipid-associated nerve macrophages in #polyneuropathies.
Explore: pns-atlas.mzhlab.com
A tweetorial 1/13
doi.org/10.1038/s414...

Our paper was just published in Nat Comm. First single‑nucleus atlas of human sural nerves in health and disease. Identified lipid‑phagocytosing macrophages and focal perineurial hyperplasia especially in immune-mediated polyneuropathies. rdcu.be/eB2Dg
23.08.2025 12:25 — 👍 5 🔁 3 💬 0 📌 0