Being in the same tweet as Olke Uhlenbeck is a true honor. Thank you @jhdcate.bsky.social
12.12.2024 22:45 — 👍 1 🔁 0 💬 0 📌 0
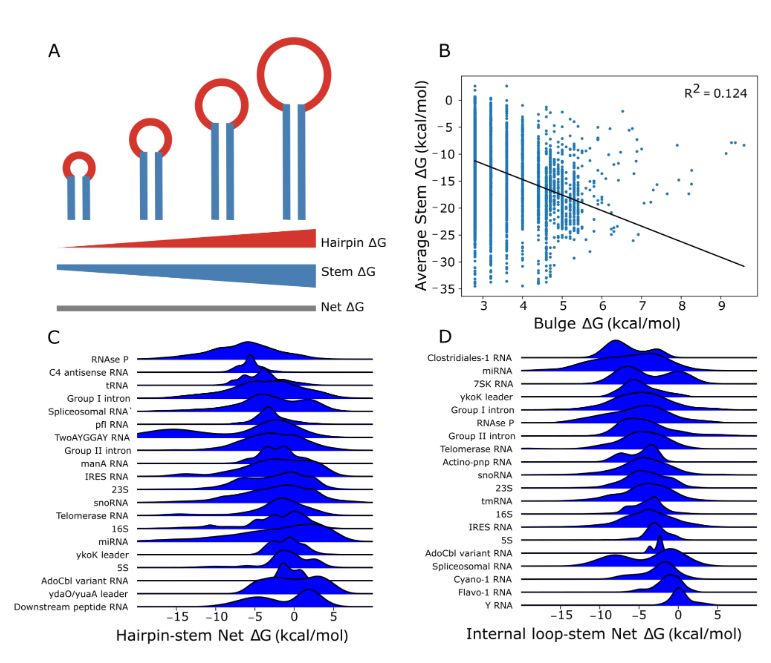
Excited to announce a new collaborative preprint about a structural concept 'local stability compensation'. This states structurally important motifs must be flanked by more stable helices. We observe this effect natural occurring RNAs and experimentally evaluate it. www.biorxiv.org/content/10.1...
12.12.2024 18:19 — 👍 17 🔁 5 💬 0 📌 1
Hi @incarnatolab.bsky.social thanks very much. Yes we ran this already and are processing now. There are many interesting things to checkout.
29.11.2024 20:09 — 👍 1 🔁 0 💬 0 📌 0
@rodrigo-reis.bsky.social We are doing that right now. I totally agree.
25.11.2024 19:47 — 👍 2 🔁 0 💬 0 📌 0
@gallardo-seq.bsky.social yes, I absolutely agree.
25.11.2024 18:25 — 👍 0 🔁 0 💬 0 📌 0
Our results provide a quantitative framework for interpreting DMS reactivity patterns in RNA. This enables more sophisticated structure prediction algorithms that consider local sequence context, non-canonical interactions, and three-dimensional features - moving beyond simple base-pair predictions.
25.11.2024 15:52 — 👍 0 🔁 0 💬 0 📌 0
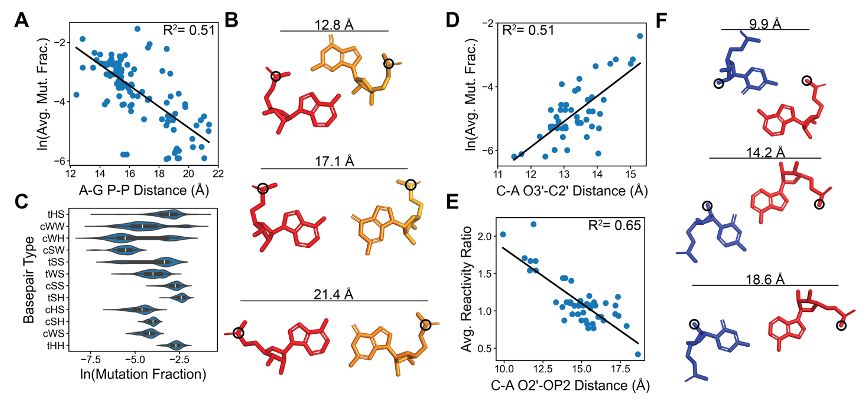
Most significantly, we discover that DMS reactivity correlates strongly with atomic distances in non-canonical base pairs. These quantitative relationships demonstrate that DMS chemical mapping data encodes detailed information about RNA 3D structure.
25.11.2024 15:52 — 👍 2 🔁 0 💬 1 📌 0
Our results provide a quantitative framework for interpreting DMS reactivity patterns in RNA. This enables more sophisticated structure prediction algorithms that consider local sequence context, non-canonical interactions, and three-dimensional features - moving beyond simple base-pair predictions.
25.11.2024 15:51 — 👍 1 🔁 0 💬 0 📌 0
Our results provide a quantitative framework for interpreting DMS reactivity patterns in RNA. This enables more sophisticated structure prediction algorithms that consider local sequence context, non-canonical interactions, and three-dimensional features - moving beyond simple base-pair predictions.
25.11.2024 15:51 — 👍 0 🔁 0 💬 0 📌 0

Most significantly, we discover that DMS reactivity correlates strongly with atomic distances in non-canonical base pairs. These quantitative relationships demonstrate that DMS chemical mapping data encodes detailed information about RNA 3D structure.
25.11.2024 15:51 — 👍 0 🔁 0 💬 0 📌 0
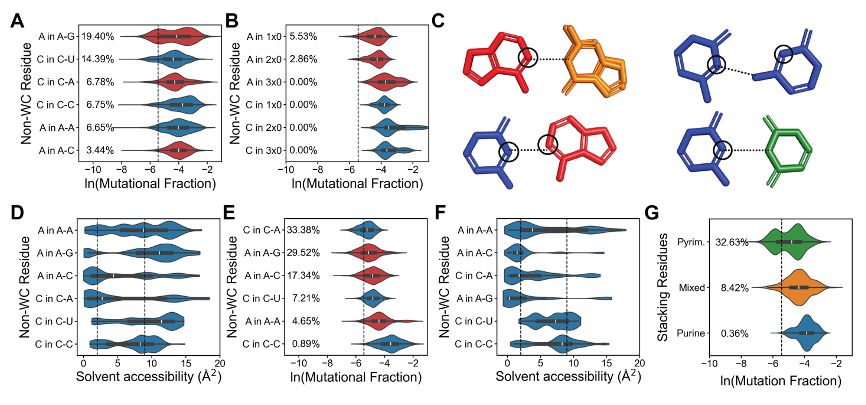
We find that 11% of non-Watson-Crick nucleotides show protection from DMS similar to Watson-Crick pairs. This protection stems from hydrogen bonding and reduced solvent accessibility. Sequence context can alter reactivity up to 100-fold in specific non-canonical pairs.
25.11.2024 15:50 — 👍 0 🔁 0 💬 1 📌 0
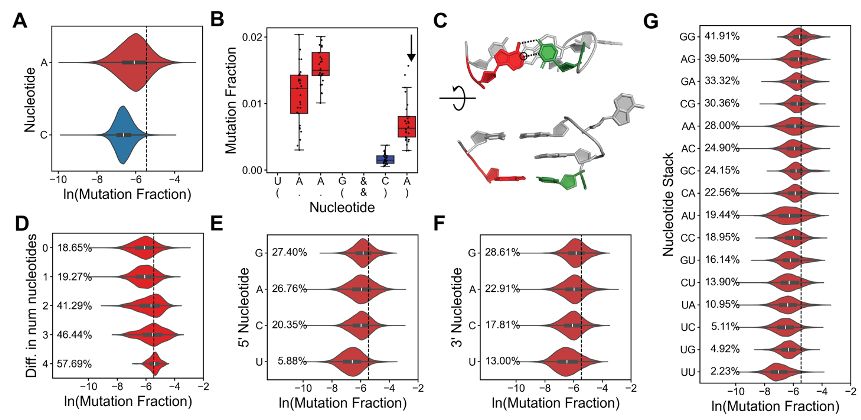
We analyzed flanking WC pairs and found structural features that determine their DMS reactivity. A-U pairs are 19-fold more reactive than G-C pairs, purine neighbors increase reactivity, and junction asymmetry correlates with higher reactivity.
25.11.2024 15:49 — 👍 1 🔁 0 💬 4 📌 0
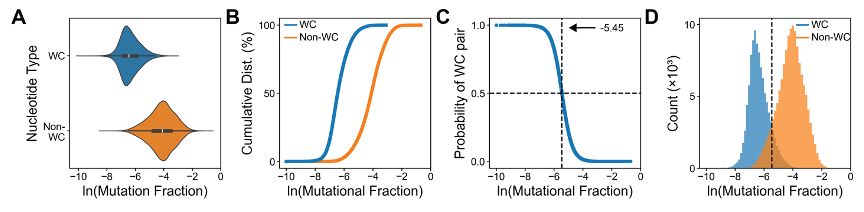
Analysis of our comprehensive dataset reveals DMS reactivity exists on a continuous spectrum rather than discrete states. We observe ~10% overlap between Watson-Crick and non-Watson-Crick nucleotides, demonstrating that simple reactivity thresholds cannot reliably determine base-pairing status.
25.11.2024 15:46 — 👍 2 🔁 0 💬 1 📌 0
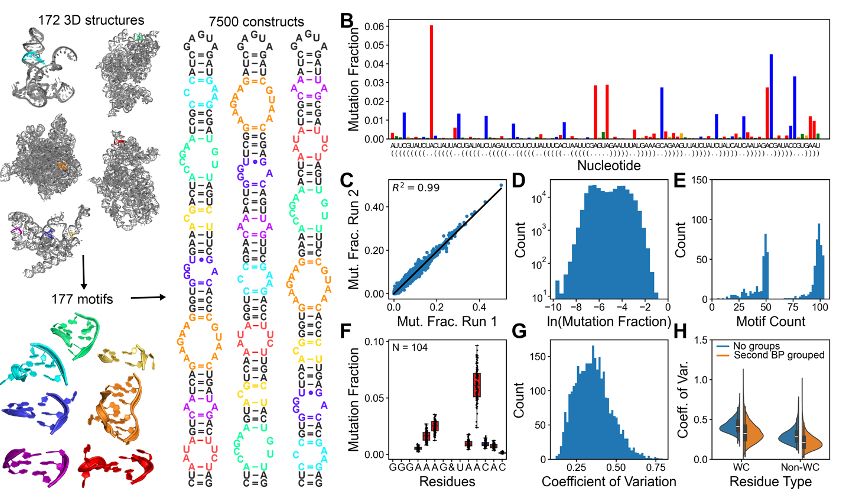
To correlate DMS reactivity with RNA structure, we built a massive library of 7,500 RNA constructs containing multiple junctions with known 3D structures. Our measurements are highly reproducible (R²=0.99), span four orders of magnitude, and reveal that RNA motifs have unique DMS profiles.
25.11.2024 15:45 — 👍 3 🔁 1 💬 1 📌 0

A quantitative framework for structural interpretation of DMS reactivity
Dimethyl sulfate (DMS) chemical mapping is widely used for probing RNA structure, with low reactivity interpreted as Watson-Crick (WC) base pairs and high reactivity as unpaired nucleotides. Despite i...
🧬 Excited to share our new preprint! DMS chemical mapping, a key technique for studying RNA structure. Everyone assumes low DMS reactivity = Watson-Crick , high = non-WC. However, analyzing 7,500 RNA structures containing known 3D structures reveals it's not that simple. doi.org/10.1101/2024...
25.11.2024 15:42 — 👍 52 🔁 22 💬 5 📌 3
@incarnatolab.bsky.social Same. Thanks for the shoutout. It's nice to see you here.
22.11.2024 15:32 — 👍 1 🔁 0 💬 0 📌 0
I hope this place is more positive and science-friendly than Twitter. I look forward to chatting about awesome science.
19.11.2024 18:31 — 👍 5 🔁 0 💬 2 📌 0
Our lab seeks an agile and predictive understanding of how RNAs structurally code for information processing and replication in living systems.
Associate Professor of Biophysics and Chemistry @Umich. Structural Biology. NMR. Biochemistry.
Chemical Biologist. Tenure track assistant prof @EPFL. Trying to make sense of the world by changing one variable at a time. https://www.epfl.ch/labs/libn/
The Royal Society of Chemistry Nucleic Acids Interest Group. Bringing together everyone who loves nucleic acids❤️🧬
Science integrity consultant and crowdfunded volunteer, PhD.
Ex-Stanford University. Maddox Prize/Einstein F Award winner
NL/USA/SFO.
#ImageForensics
@MicrobiomDigest on X.
Blog: ScienceIntegrityDigest.com
Support me: https://www.patreon.com/elisabethbik
Assistant professor at Baylor College of Medicine: Therapeutic Innovation Center and Department of Biochemistry and Molecular Pharmacology.
Tackling RNA regulation and structure with computation and experiments.
My views are my own.
RNA and ciliate molecular biologist, college prof, mom, living and working in the Pacific NW 👩🏻🔬🧬👩🏻🏫🔬
https://biology.wwu.edu/people/lees65
RNA and DNA geek | Structure-function of nucleic acids | Assistant Professor at the University of Florida | My views. Website: https://sites.google.com/view/passalacqualab/
Studying genomics, machine learning, and fruit. My code is like our genomes -- most of it is junk.
Assistant Professor UMass Chan, Board of Directors NumFOCUS
Previously IMP Vienna, Stanford Genetics, UW CSE.
Exploring RNA & RBPs in cancer: from cell edges to the chromatin. Microscopy lover, proteomics newbie. Likes computer sciences, physics & photography. Also an illustrator and graphic designer for Science and Biology (see: @STEMDORADOscimag)
Associate Professor @YaleNeuro • co-Director of Graduate Studies @Yale_INP • @YaleRNA • RNA Neurobiology • Neurodegenerative disease • Immigrant • 🐶 dad • Alum @WhiteheadInst @HopkinsNeuro @PKU1898 🏳️🌈
Professor at Oregon State University. Computational biology and nucleic acid bioinformatics.
Organic chemist interested in the origins of life. Moranlab.com
🇨🇦 CRC1 and Prof @uottawa.ca
🇫🇷 part-time @unistra.fr
Biophysics PhD Student with DasLab & ChiuLab.
Pairing RNA structure & cryoEM to better understand both!
King's College, Science & Security '19
Harvey Mudd '18
Biochemist who enjoys many-a-things. He/Him 🇺🇸🇨🇴
RNA scientist | Assistant Prof. Aarhus University and Lundbeck Fellow | Editor Molecular Therapy Nucleic Acids
Scientist interested in structural biology, vaccine design, antibodies, protein engineering… pretty much everything