

Excited to speak at the
@stanforddata.bsky.social Sustainability Data Science conference today!
I’ll be presenting ClimateX, our work on LLM confidence calibration in the climate science domain.
📄 Paper: arxiv.org/abs/2311.17107
💻 Code: github.com/rlacombe/Cli...
10.04.2025 16:27 — 👍 1 🔁 0 💬 0 📌 0
That’s a bit of relief at least! 🐚🦪
05.04.2025 07:23 — 👍 1 🔁 0 💬 0 📌 0
Poster time! Fantastic workshop on experimental design in AI for Science today.
Paper: arxiv.org/abs/2503.17368
Workshop: edai4science.github.io
04.04.2025 05:31 — 👍 2 🔁 0 💬 0 📌 0

Nobel laureate talk time!
With David Baker at the Stanford experimental design in AI for Science today.
03.04.2025 20:31 — 👍 1 🔁 0 💬 0 📌 0
Pierre Clostermann during WWII
02.04.2025 09:51 — 👍 1 🔁 0 💬 0 📌 0
Experimental Design: AI for Science
Excited to be presenting this work at the
@stanfordbiosci.bsky.social workshop on Experimental Design in AI for Science next week!
More info on the event here: edai4science.github.io
27.03.2025 20:18 — 👍 0 🔁 0 💬 1 📌 0
These results highlight the limits of the (extremely successful) evolutionary approach to protein structure prediction.
We will likely need physics-informed models to reach accurate 3D structure predictions on rare PTMs and out-of-domain proteins beyond evolutionary priors.
27.03.2025 20:18 — 👍 0 🔁 0 💬 1 📌 0
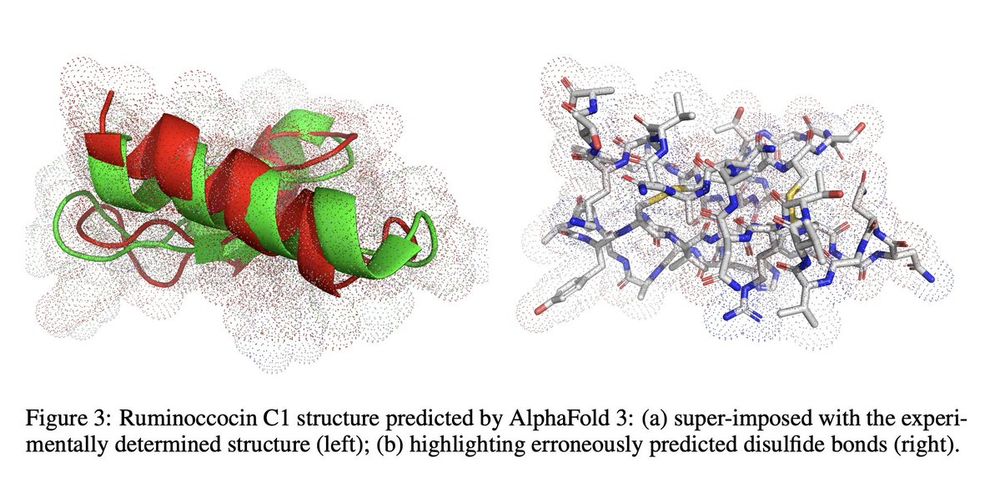
No model achieved crosslink distances near the experimentally observed 1.8Å sulfur-to-α-carbon bond length, with average performance at 10-12Å.
Most models incorrectly predicted disulfide bonds instead of thioether crosslinks, highlighting their reliance on evolutionary priors.
27.03.2025 20:18 — 👍 0 🔁 0 💬 1 📌 0
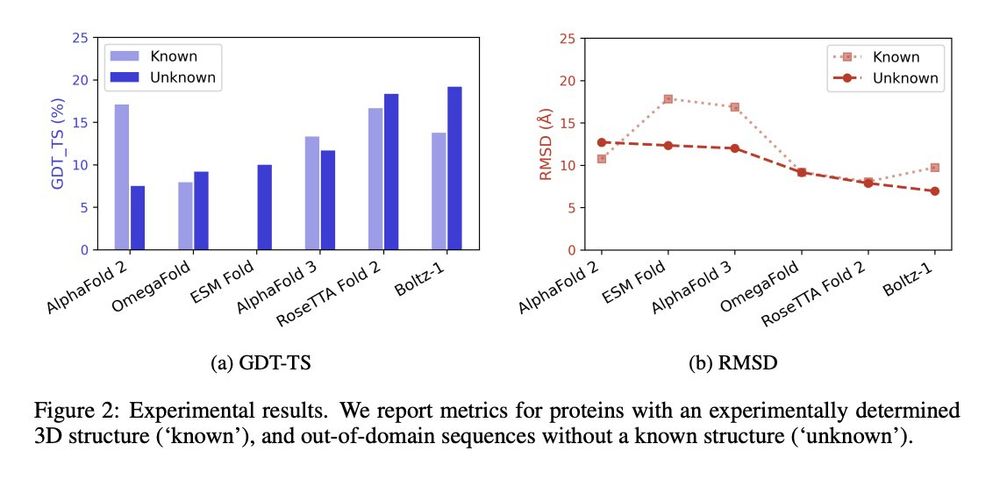
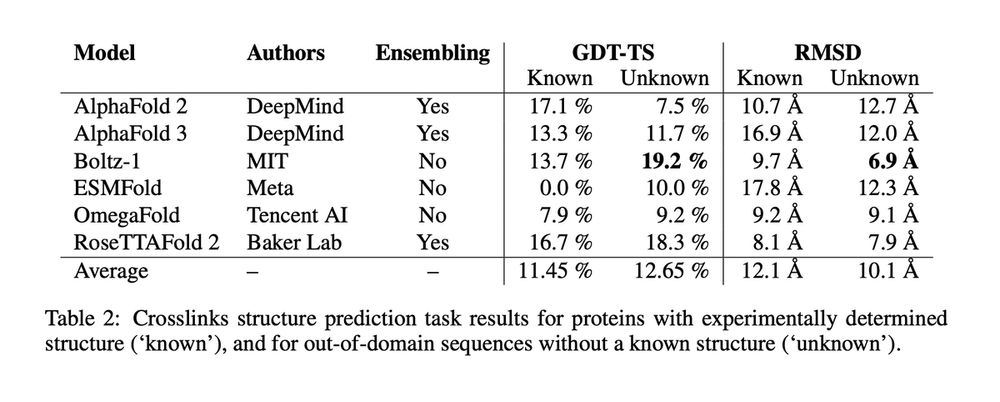
We evaluate 6 leading protein structure predictors—AlphaFold 2 and 3, Boltz-1, ESMFold, OmegaFold, and RoseTTAFold 2—on the 10 known sactipeptides.
All models exhibit limited performance, with an average GDT-TS of only 11.5% for known sactipeptides and 12.6% for unknown ones.
27.03.2025 20:18 — 👍 0 🔁 0 💬 1 📌 0
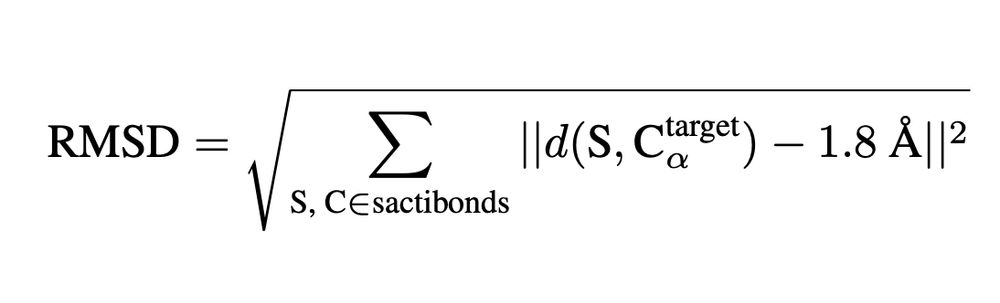
This helps us probe how deep learning models generalize beyond evolutionary priors.
We introduce a new, zero-shot benchmark for structure models: measuring how the 3D conformations predicted for sactipeptides match the geometry imposed by their known thioether crosslinks.
27.03.2025 20:18 — 👍 0 🔁 0 💬 1 📌 0
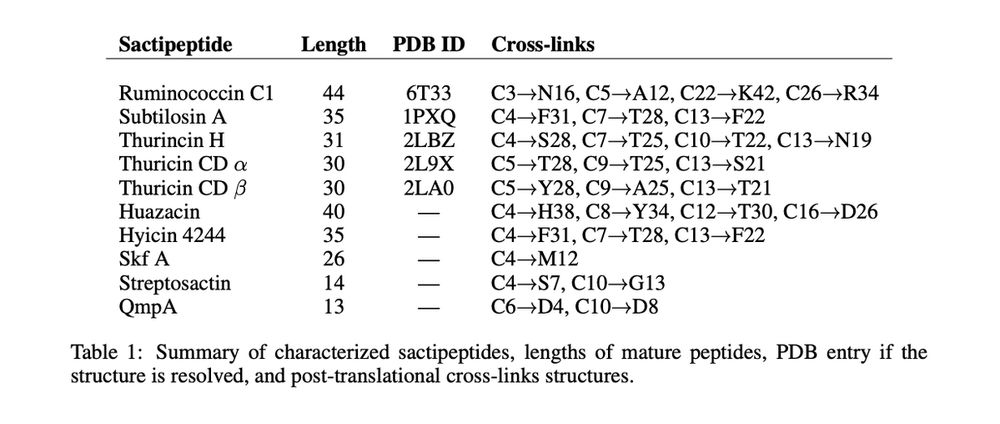
These non-canonical crosslinks are challenging and underrepresented in structural datasets. Crucially, *only 5* of the 10 known sactipeptides have a resolved 3d conformation with a PDB entry. *But* their 2d crosslink structure is known, which constrains possible 3d geometries!
27.03.2025 20:18 — 👍 0 🔁 0 💬 1 📌 0
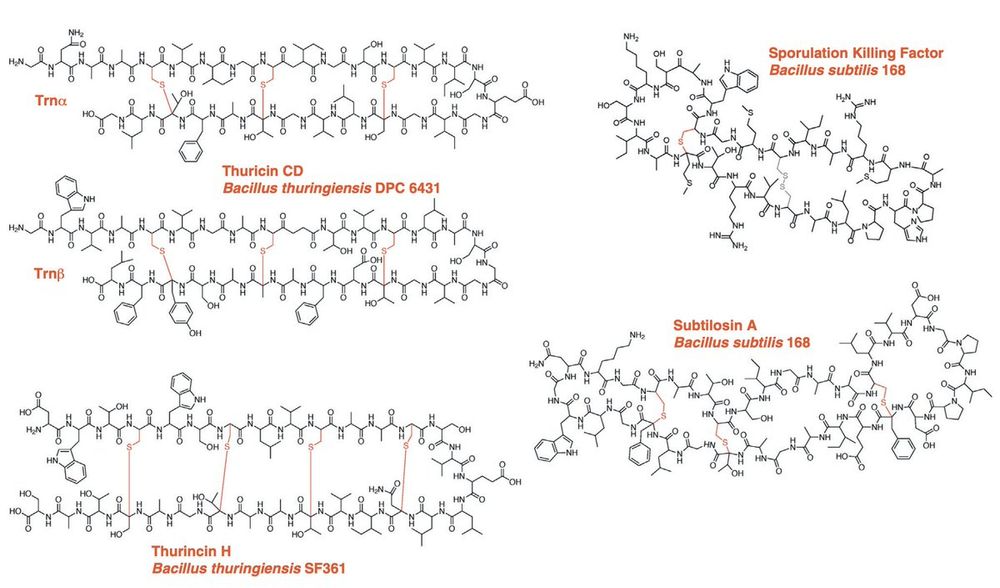
Evolution-based protein structure prediction models have revolutionized structural biology but struggle with rare post-translational modifications (PTMs). We evaluate them on sactipeptides, a rare class of 10 known peptides with unique sulfur-to-α-carbon thioether crosslinks.
27.03.2025 20:18 — 👍 0 🔁 0 💬 1 📌 0
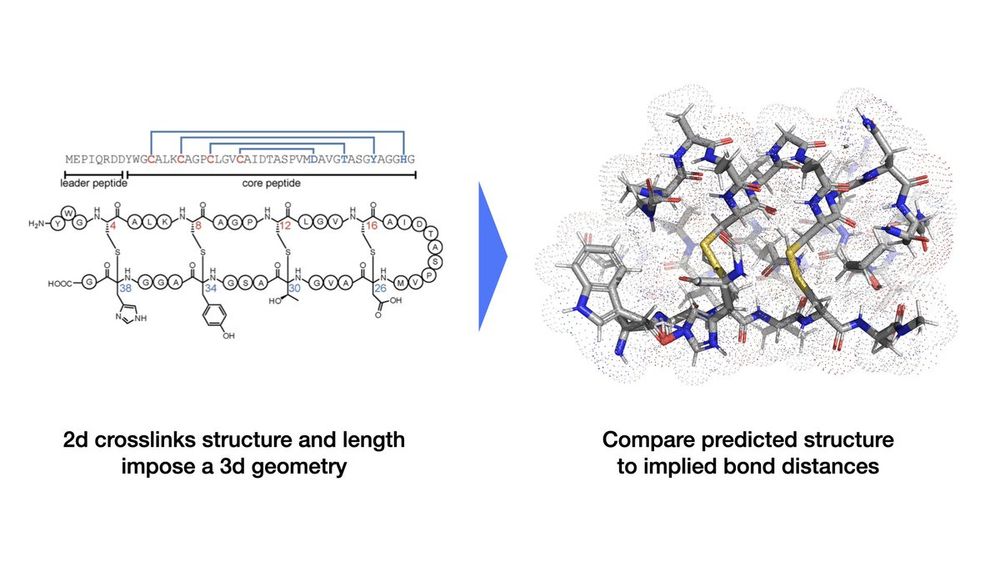
🔔🔔 New bioML paper alert! 🔔🔔
We evaluate evolutionary protein structure prediction models using the geometric structure of non-canonical crosslinks from a rare class of post-translationally modified peptides.
A thread. 🧵
27.03.2025 20:18 — 👍 1 🔁 0 💬 1 📌 0
"But you're so much more handsome than that!"
– my beloved wife, when shown a Ghiblified selfie of ourselves.
Right answer!! I'm swooning! 🥰🥰🥰
27.03.2025 18:39 — 👍 2 🔁 0 💬 0 📌 0
Pre-ordered! 🛍️🤞
25.03.2025 18:00 — 👍 0 🔁 0 💬 0 📌 0
This is much deeper than it sounds at first. Absolute nugget of wisdom.
25.03.2025 17:51 — 👍 0 🔁 0 💬 0 📌 0
Oooh that looks beautiful! When can we pre-order?
25.03.2025 17:26 — 👍 0 🔁 0 💬 1 📌 0
Anecdotally, @dcrainmaker.com has a treasure trove of GPS accuracy tests in land and marine conditions for most watches and trackers on the market.
25.03.2025 01:04 — 👍 2 🔁 0 💬 0 📌 0
LLMs are Farenheit 451 for digitized human knowledge.
There’s something comforting knowing that the ghost of every written word will live on, for as long as we can preserve open weights, and the GPUs to bring them to life.
22.02.2025 07:39 — 👍 1 🔁 0 💬 0 📌 0
Chemical engineering rules everything around us. ⚗️
AI 🤖, bio 🧬, chips 💻, energy 🔋, fertilizers 🚜, materials ⛏️, space 🛰️ ...
All the technologies shaping the world sit downstream from chemistry fundamentals. 👩🔬
Hold on to your periodic table and let's go! 🚀
04.01.2025 03:06 — 👍 8 🔁 0 💬 0 📌 0

Few things are more heart wrenching than a walk by a wild, pristine, remote beach at the confines of civilization…
…only to find billions of tiny speckles of microplastics all over the sand, which we *know* will end up in our bodies by way of the food chain.
Very sad.
23.12.2024 01:59 — 👍 10 🔁 0 💬 0 📌 0
This guy is *such* a jerk. Ugh! Only good thing is it’s out in the open for anyone to see.
15.12.2024 07:31 — 👍 6 🔁 0 💬 0 📌 0

1 failure per 1000 GPU•days is the going hardware MTF rate for large training runs these days.
#NeurIPS2024
12.12.2024 00:50 — 👍 0 🔁 0 💬 0 📌 0

Craziest talk of the day so far: you can recognize which room someone stands in with embeddings of firing rate maps of their place cells neurons!
#NeurIPS2024
11.12.2024 18:53 — 👍 1 🔁 0 💬 0 📌 0

Deep learning is chemical engineering for information: layers are process units, diffusion is transport, entropy is the loss function…
Don’t believe me? Then why is the NeurIPS opening talk literally starting with the Haber Bosch process!?
11.12.2024 18:46 — 👍 3 🔁 0 💬 0 📌 0

Made it to #NeurIPS2024!
Here until Sunday, ping me for coffee, a walk and talk, or some Tim Hortons maple glazed donuts! 🇨🇦
11.12.2024 03:43 — 👍 2 🔁 0 💬 0 📌 0

I used to add little incandescent light bulbs in my notes, for highlights.💡
But it’s time to decarbonize. 🔋
So now I add little LEDs! ⚡️
08.12.2024 23:44 — 👍 3 🔁 0 💬 0 📌 0

We can just fix things.
03.12.2024 05:49 — 👍 3 🔁 0 💬 0 📌 0
I am a good bot posting excerpts from the seminal text by W.S. Broecker and T-H Peng for you. Many oceanographers started their journey of ocean discovery with this text
Husband, dad, veteran, writer, and proud Midwesterner. 19th US Secretary of Transportation and former Mayor of South Bend.
Virtual Chemist; Liúdramán; Personal Opinions
Group leader at Institut Jacques Monod. When I say "epigenetics" what I really mean is "DNA methylation".
www.maximgreenberglab.com
Dad, husband, President, citizen. barackobama.com
Google Chief Scientist, Gemini Lead. Opinions stated here are my own, not those of Google. Gemini, TensorFlow, MapReduce, Bigtable, Spanner, ML things, ...
Computational Biophysicist, Amateur Photographer, History+Language Nerd.
All views my own.
Website: https://martinvoegele.github.io/
DGA PublicSpace & Facilities #Paris17
Radio facilitator Parlez-moi d'IA sur @radiocausecommune.bsky.social (Open...Source, #Data, #IA, Innov & Gov) My own opinion #Velotaf #Clichy
https://linktr.ee/AgentNuM
Product Marketing Lead @NVIDIA
| PhD @UMBaltimore | omics, immuno/micro, AI/ML | 🇺🇸🇸🇰 |
Posts are my own views, not those of my employer.
🧪 Research Scientist @nvidia and PhD student @Oxford staring at proteins all-day
🧑💻 Website/Blog: https://kdidi.netlify.app/
🤖 GitHub: https://github.com/kierandidi
📚 Prev. Cambridge/Heidelberg
🏳️🌈 NVIDIA & Duke. Was Allianz, VantAI, TUM. BioCS+ML dude.
Lab page: https://machine.learning.bio
GScholar: https://scholar.google.com/citations?user=4q0fNGAAAAAJ
Computational biologist. RWTH Aachen, Stanford, U of Cambridge. ex-AZ, ex-Bayer. NVIDIA
AI PhD Student at University of Oxford, Applied Research Scientist at NVIDIA
HCLS @ NVIDIA. Single mom by choice. Views are my own. (she/her)
Digital Biology @ NVIDIA
Senior AI applied scientist for Drug Discovery
https://research.nvidia.com/labs/genair/
MIT PhD Student - ML for biomolecules - https://hannes-stark.com/



















