Open licenses power discovery!
Melissa Harrison shares a @pfamdb.bsky.social use case:
Pfam uses AI to extract information about known protein domains
This helps predict protein function & identify potential drug targets! 💊
#ChooseOpenWebinar for live updates 💡
#MedLib #PhDsky #OpenAccess
09.07.2025 16:11 — 👍 0 🔁 2 💬 0 📌 0

Poster A-098: Pfam database in 2025 – towards expanding protein families coverage
Nicole Morveli Flores
Session A: 21/07/2025
10:00 – 11:20 and
16:00 – 16:40 BST
Image credit: BioImage Archive accession S-BSST584.
Design credit: Karen Arnott/EMBL-EBI
InterProScan 6: a modern large-scale protein function annotation pipeline
Matthias Blum
21/07/2025 14:20
Room 03A
Image credit: BioImage Archive accession S-BSST584. Design credit: Karen Arnott/EMBL-EBI

Poster A-205: Carbohydrate active enzymes in Pectobacteriaceae: coevolving enzyme sets and host adaptation
Emma Hobbs
Session A: 21/07/2025
10:00 – 11:20 and
16:00 – 16:40 BST
Image credit: BioImage Archive accession S-BSST584.
Design credit: Karen Arnott/EMBL-EBI

Accelerating protein family classification in InterPro with AI innovations
Matthias Blum
22/07/2025 16:40
Room 12
Image credit: BioImage Archive accession S-BSST584.
Design credit: Karen Arnott/EMBL-EBI
🧬 The InterPro/Pfam team will be presenting at #ISMB2025 next week in Liverpool!
🎤 Multiple talks & 📊 posters on protein analysis & classification
💻🧪
18.07.2025 14:11 — 👍 1 🔁 1 💬 0 📌 0

The InterPro/Pfam team members attending ISMB/ECCB 2025: Alex Bateman, Emma Hobbs, Matthias Blum, Nicole Morvelli, Antonina Andreeva
Image credit: BioImage Archive accession S-BSST584.
Design credit: Karen Arnott/EMBL-EBI
🧬 The InterPro/Pfam team will be presenting at #ISMB2025 next week in Liverpool!
Join us July 20-24 at the world's largest #bioinformatics conference!
#ISMBECCB2025 #ProteinAnalysis #ComputationalBiology
💻🧪
18.07.2025 14:09 — 👍 3 🔁 1 💬 0 📌 0
Come chat to us at @jobim2025.bsky.social!
07.07.2025 08:24 — 👍 1 🔁 0 💬 0 📌 0

Promotional slide for a webinar titled "What happens when you choose closed? What Creative Commons licenses mean for research and AI." The slide features photos and names of five speakers: Melissa Harrison (Europe PMC / EMBL-EBI), Katherine Hughes (EMBL), Taylor Campbell (Creative Commons), Monica Granados (Creative Commons), and Kat Walsh (Creative Commons). Logos for Europe PMC, Creative Commons, and EMBL are shown at the bottom. The background is green with white text.
Can AI train on my openly licensed research? Do I retain copyright if I choose a CC BY license? Why can’t I reuse a figure from my own paper?
Get the answers in a webinar with Creative Commons, #EuropePMC & EMBL
🗓️ 9 Jul
⏰ 15:00 UTC
🔗 embl-org.zoom.us/webinar/regi...
#OpenScience #AcademicSky 🧪
23.06.2025 09:02 — 👍 12 🔁 9 💬 0 📌 1

Pfam 37.4
318 new families
4 new clans

Content
Over 24,736 entries describing protein families, domains and repeats.
🚀 HUGE NEWS: Pfam 37.4 is LIVE!
318 brand-new protein families + 4 new clans! 🧬
Total collection now: 24,736 entries! 📈
Get exploring via InterPro website, API & Pfam FTP!
💻🧪
20.06.2025 08:47 — 👍 10 🔁 1 💬 0 📌 0

InterPro 106.0
19th June 2025
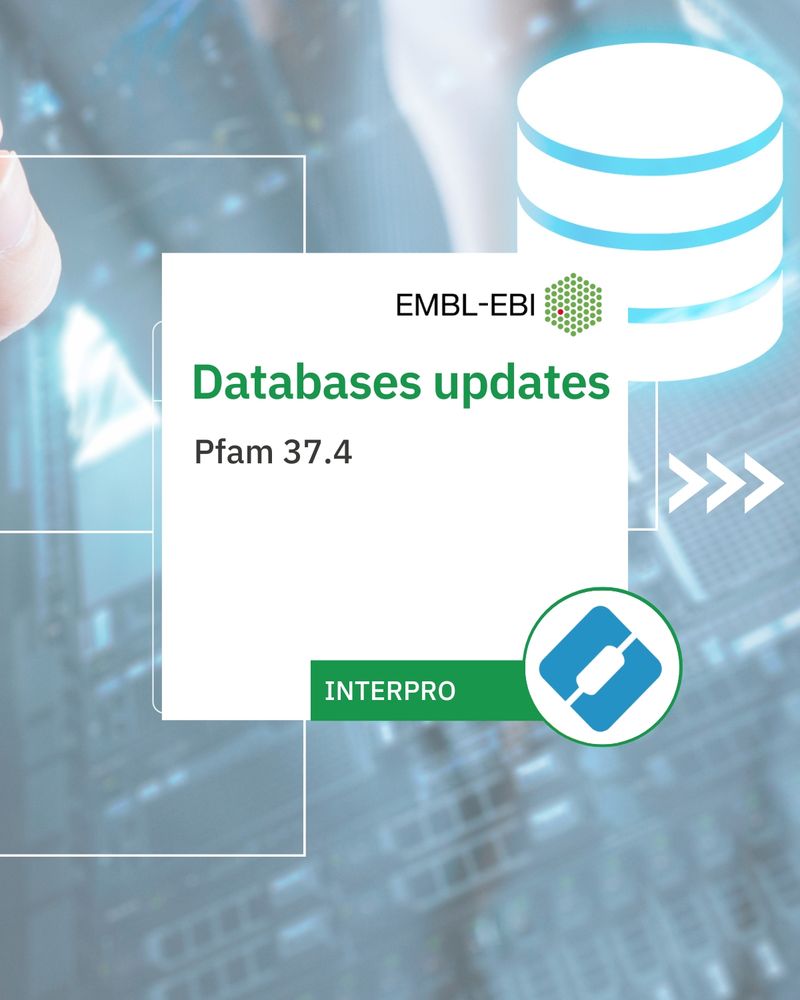
Databases updates:
Pfam 37.4

New entries:
683 new entries with:
613 Pfam
78 NCBIfam
30 CDD
4 PANTHER
...
🚀 InterPro 106.0 is now live!
✨ Updated to Pfam 37.4
✨ 683 new entries added
✨ Enhanced protein classification coverage
Explore the latest annotations and discover new protein insights.
Explore it now: interpro.ebi.ac.uk 💻🧪🧬
#Bioinformatics #ProteinScience #Pfam #EMBLEBI
20.06.2025 08:22 — 👍 8 🔁 2 💬 0 📌 0
What’s new in the redesigned PDBe entry pages?
🧪 Cleaner layout
🧪 Interactive 3D viewers
🧪 Contextual annotations
🧪 Easier navigation
See for yourself:
wwwdev.ebi.ac.uk/pdbe/entry/p...
#PDBeBeta #Bioinformatics #Biochemistry #Science
16.06.2025 15:40 — 👍 11 🔁 5 💬 0 📌 0
🧬 MAJOR UPDATE: InterPro-N matches now available in InterPro!
1.8 billion AI-driven protein annotations developed with Google DeepMind are now live.
✨ Look for the sparkles icon to identify them
📊 Options menu lets you view: InterPro only | InterPro-N-only | Stacked
💻🧪
15.05.2025 17:07 — 👍 4 🔁 1 💬 0 📌 0
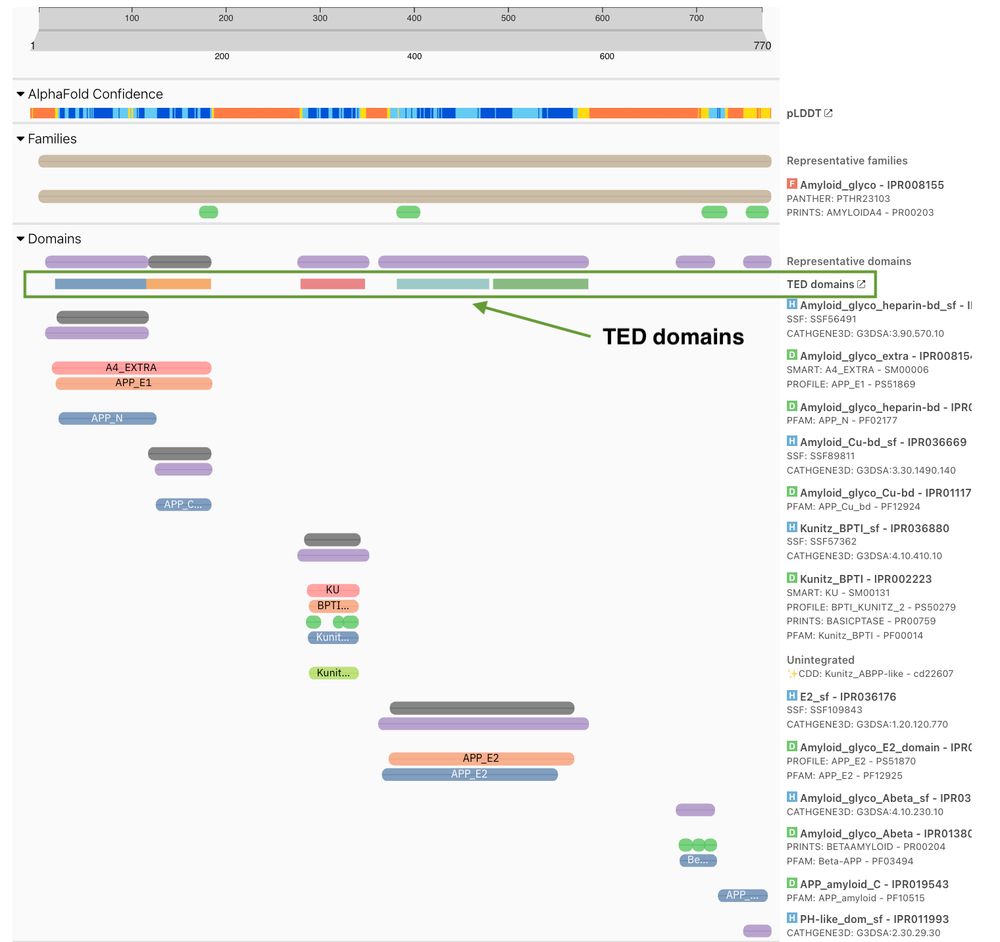
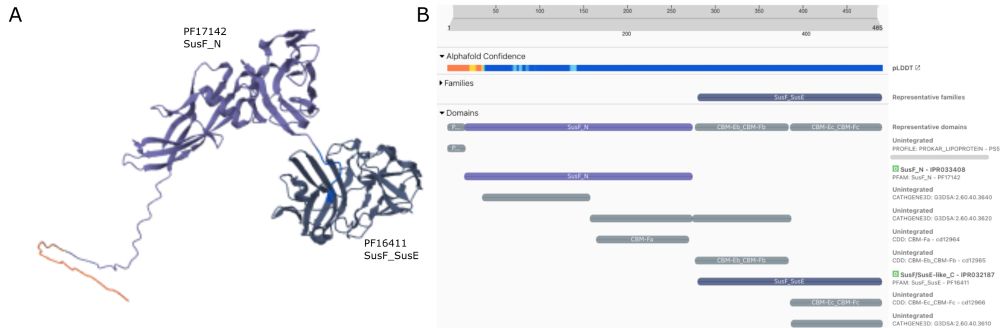
TED domains alongside InterPro annotations for UniProt P05067
🔍 NEW: TED domains in InterPro 105.0!
For the first time, see AlphaFold-derived structural domains directly alongside InterPro annotations in one powerful visualisation. Bridging the sequence-structure divide in protein analysis. 💻🧪
Explore now: www.ebi.ac.uk/interpro/pro...
08.05.2025 14:54 — 👍 13 🔁 5 💬 0 📌 0
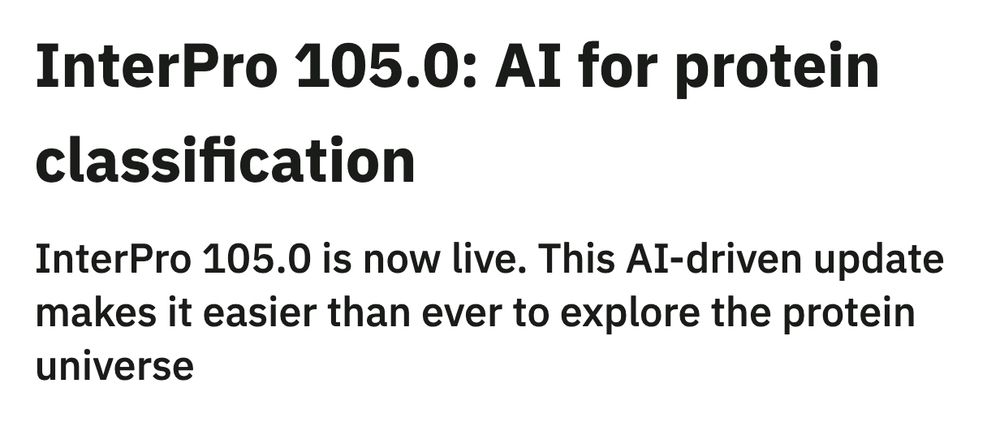
InterPro 105.0: AI for protein classification
InterPro 105.0 is now live. This AI-driven update makes it easier than ever to explore the protein universe
📝 Check out our new blog post highlighting the groundbreaking features in InterPro 105.0!
Learn how InterPro-N, TED domains and viral protein structures are transforming protein classification: ebi.ac.uk/about/news/u...
01.05.2025 16:35 — 👍 3 🔁 1 💬 0 📌 0

🔥 Pfam 37.3 just dropped! 🚀
353 incredible new protein families + 7 new clans! 🧬✨
Revolutionise your research NOW via @interprodb.bsky.social website, API & Pfam FTP!
25.04.2025 08:24 — 👍 1 🔁 1 💬 0 📌 0

InterPro 105.0
24th April 2025

Databases updates
Pfam 37.3
PROSITE patterns 2025_01
PROSITE profiles 2025_01
HAMAP 2025_01

New entries
342 new entries with:
318 Pfam
40 NCBIfam
12 Prosite profiles
6 PANTHER
5 CDD

New features
InterPro-N annotations
BFVD structure predictions
Simplified structure pages
📢InterPro release 105.0 is live!
✨Update to Pfam 37.3, PROSITE patterns 2025_01
PROSITE profiles 2025_01 and HAMAP 2025_01
✨342 new entries
✨Addition of InterPro-N annotations
✨Addition of BFVD structure predictions from @martinsteinegger.bsky.social's lab
Have a look: ebi.ac.uk/interpro/htt...
💻🧪🆕
24.04.2025 16:07 — 👍 19 🔁 10 💬 1 📌 0
Want to use the InterPro API but not sure how to start? Join this upcoming workshop, where @typhainepl.bsky.social will guide you through the fundamentals and practical applications. Discover how to integrate this powerful tool into your workflow! Register now to secure your spot. 💻🧪
02.04.2025 09:36 — 👍 4 🔁 2 💬 0 📌 0

InterPro 104.0
6th February 2025
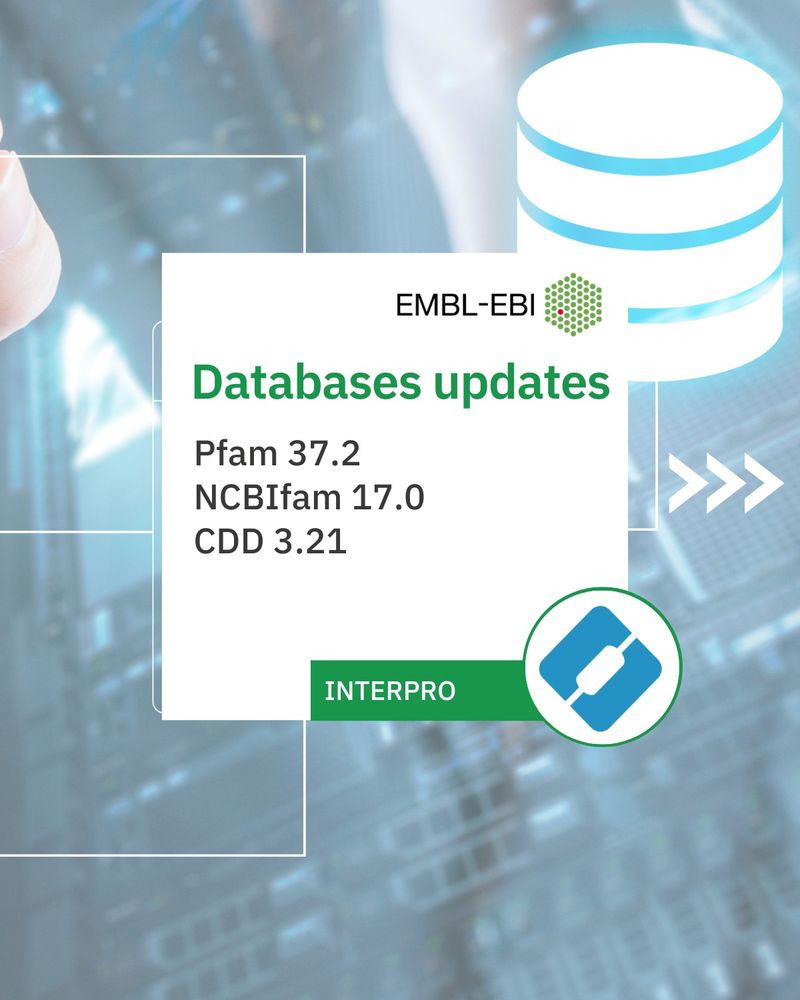
Databases udpates:
Pfam 37.2, NCBIfam 17.0, CDD 3.21

661 new entries, containing:
663 Pfam
5 NCBIfam
5 PANTHER
9 CDD

New features:
Redesigned protein viewer for sequence searches
Wikipedia articles for Pfam clans
Addition of PTM from the PRIDE database
🔈InterPro release 104.0 is live!
✨Update to Pfam 37.2, NCBIfam 17.0, CDD 3.21.
✨661 new entries, including 682 signatures from the Pfam (663), NCBIfam (5), PANTHER (5), CDD (9) databases. Have a look: ebi.ac.uk/interpro/ 💻🧪
06.02.2025 14:19 — 👍 3 🔁 4 💬 0 📌 0

Pfam 37.2: 302 new families and 18 new clans
Pfam 37.2 is out 🎉It contains 302 new families and 18 new clans You can have a look at them in InterPro: ebi.ac.uk/interpro/ent... or download the release from the Pfam ftp: ftp.ebi.ac.uk/pub/database...
06.02.2025 14:46 — 👍 2 🔁 0 💬 0 📌 0
InterPro website (home of Pfam) Help page
🔍 Lost in Pfam data? Our redesigned Help page has you covered! From quick answers to deep dives into protein families, find exactly what you need at www.ebi.ac.uk/interpro/help/ 💻🧪
24.01.2025 11:04 — 👍 5 🔁 0 💬 0 📌 0
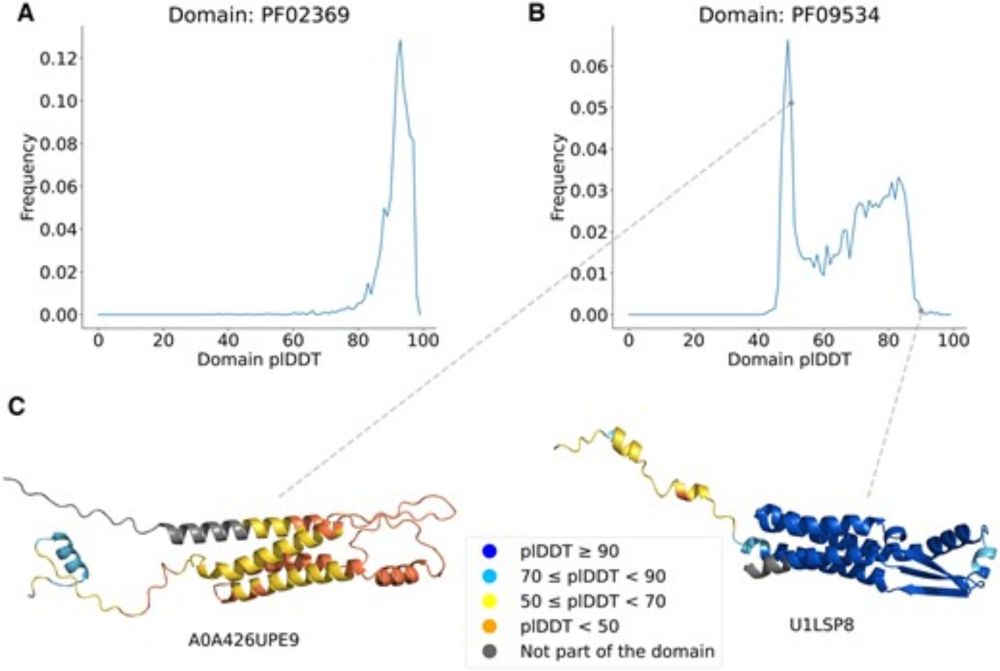
Protein structure prediction from the AlphaFold Database
Prediction confidence scores help #AlphaFold users gauge the reliability of protein structure predictions. 🖥️🧬
But things get more challenging for protein families.
A new method helps improve low confidence predictions within protein families.
academic.oup.com/bioinformati...
16.12.2024 09:28 — 👍 17 🔁 7 💬 0 📌 0
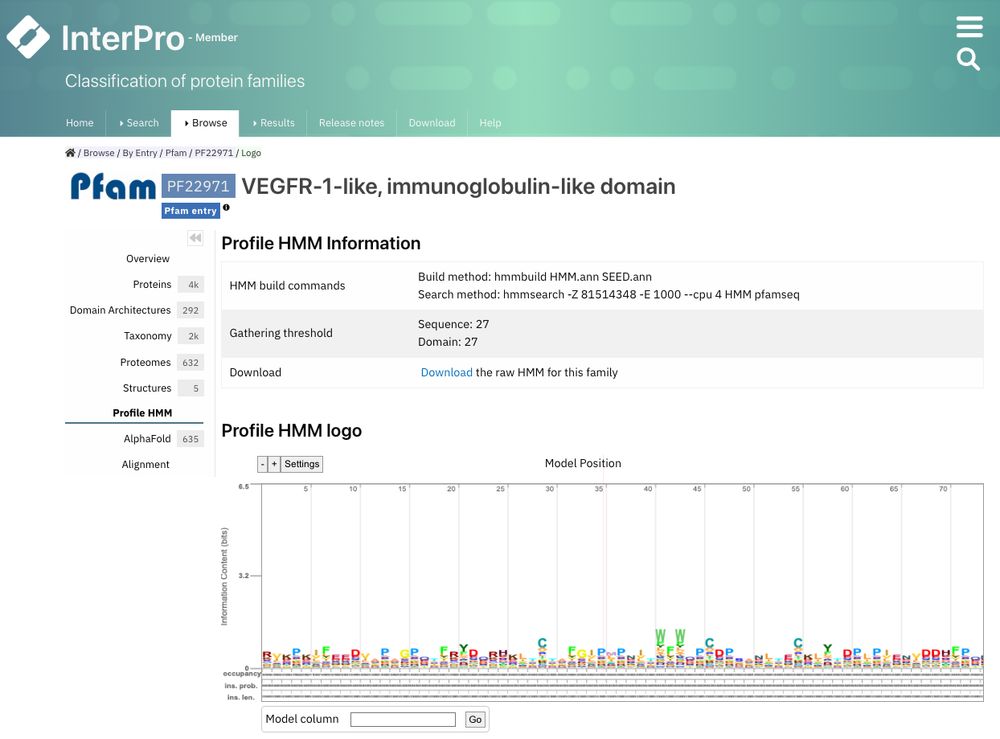
Example of Profile HMM page for Pfam PF22971. It contains HMM data and Profile HMM logo
🆕The member database signature pages have undergone a thoughtful reorganisation to improve information accessibility.
1) Curation data are now in Overview
2) HMM information and logo are now in Profile HMM
Have a look: www.ebi.ac.uk/interpro/ent...
16.01.2025 11:39 — 👍 1 🔁 2 💬 0 📌 0
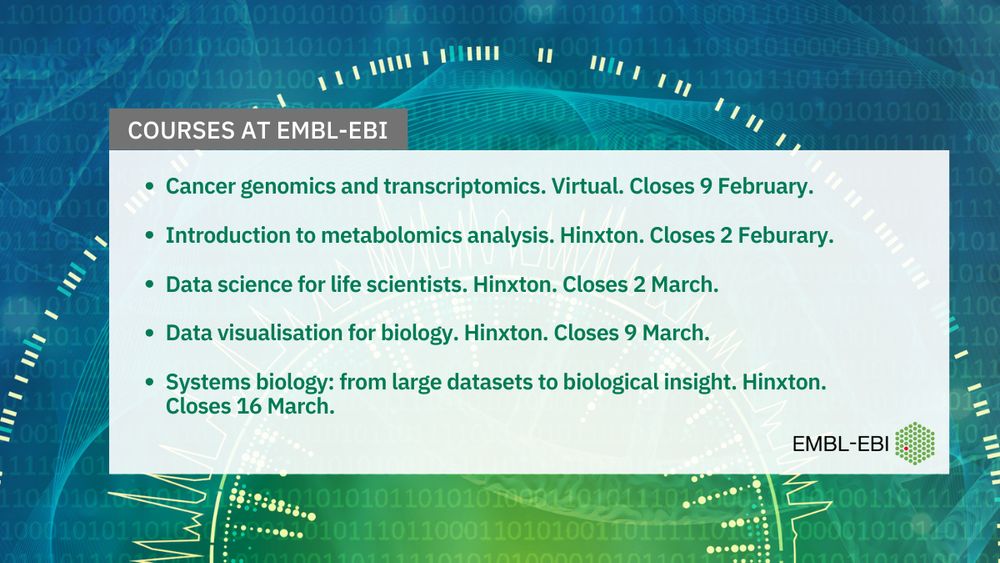
Cancer genomics and transcriptomics. Virtual. Closes 9 February.
Introduction to metabolomics analysis. Hinxton. Closes 2 Feburary.
Data science for life scientists. Hinxton. Closes 2 March.
Data visualisation for biology. Hinxton. Closes 9 March.
Systems biology: from large datasets to biological insight. Hinxton. Closes 16 March.
Welcome to 2025 🎉 We are running a number of exciting courses this year, so join us to boost your #bioinformatics, #datascience and #omics skills!
These courses are currently open for applications - find out more: www.ebi.ac.uk/training/liv...
💻🧬💻🧪
06.01.2025 11:28 — 👍 8 🔁 2 💬 0 📌 0
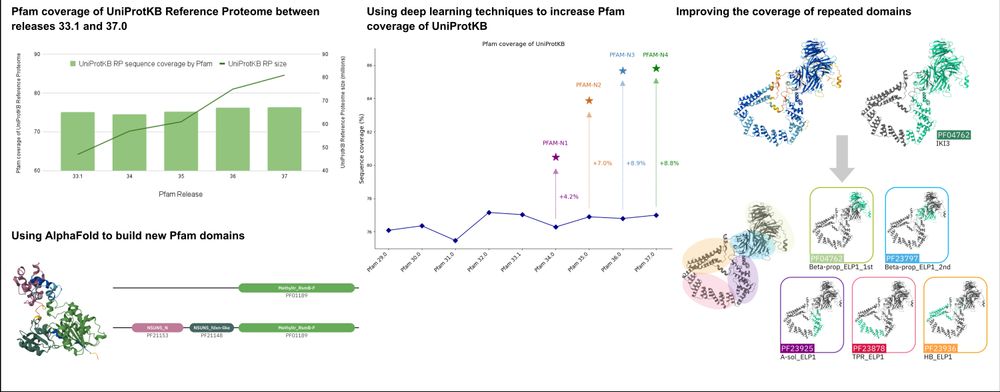
NAR graphical abstract:
Pfam coverage
Using AlphaFold to build new Pfam domains
Use of deep learning to Increase coverage of UniProtKB
Improved coverage of repeated domains
For the latest Pfam updates, have a look at our recently published paper: The Pfam protein families database: embracing AI/ML
t.co/8auCxw3mdj
29.11.2024 09:59 — 👍 3 🔁 1 💬 0 📌 1

InterPro release 103.0: 28th November 2024

Database updates:
Pfam 37.1

1029 new entries containing:
1008 Pfam
2 NCBIfam
1 PIRSF
1 PANTHER
7 CDD

New features:
Redesigned protein viewer
Improved dataset browsing
Streamlined website navigation
Member database entry page updates
InterPro release 103.0 is live! 💻🧪
✨Update to Pfam 37.1
✨1029 new entries, including 1019 signatures from the Pfam (1008), NCBIfam (2), PIRSF (1), PANTHER (1), CDD (7) databases.
Have a look: ebi.ac.uk/interpro/
29.11.2024 08:28 — 👍 5 🔁 4 💬 1 📌 1

Pfam: collection of protein families and domains
Content: Over 21,000 entries describing protein families, domains and repeats.
Training available on the EMBL-EBI training platform
We’re delighted to have joined BlueSky to engage with the scientific community about protein families, protein domains and related data. The Pfam database is a large collection of protein families, domains and repeats.
20.11.2024 10:10 — 👍 4 🔁 0 💬 1 📌 0
senior group leader @sangerinstitute. evolutionary geneticist malaria & biodiversity. lifetime member of darwin's posse. obsessive tidepooler. hearts cephalopods.
our lab leads the Malaria Cell Atlas, UK BIOSCAN, and ANOSPP projects.
We're the Crick, a biomedical research lab in London working to figure out how life works.
Home to more than 2,000 scientists and a free public exhibition space.
https://www.crick.ac.uk/
Royal Society University Research Fellow | NUAcT Fellow @ Newcastle University @mhd-newcastle.bsky.social
Investigating mycobacterial transport systems
#structuralbiology #microbiology
Instagram @kshbeckham
Synthetic biology in plants and fungi. Ag biotech, sustainability, bioeconomy. Plant hormones, lab automation, protein engineering. Crafting biology with the Biowright lab at Virginia Tech: https://sites.google.com/vt.edu/wrightlab
Chief Science Policy & Workforce Development Officer @ FASEB | Lover of shoes 👠, science 🧬, and #SciPolPony 🐴 | Devoted to Cleveland sports ⚾️🏀🏈 | Washington, DC
Chemical biologist who ❤️s protein-protein interactions, transcription factors, and undergrad education. Assistant professor at Furman Univ. Cat mom. (she/her)
Bioinformatician, Department of Biomedicine @ Uni Basel & Swiss Institute of Bioinformatics
Interested in bulk and single-cell omics, data analysis, experimental design, data visualization, R/Bioc, open science, EDI, etc 🧬 🖥️
Accelerating precision medicine through Open Data Science.
https://monarchinitiative.org/
molecular systems biology of the synapse | snsf ambizione | spatial proteomics | molecular neuroscience
https://bit.ly/MvO
Ask me about TopDown Proteomics!
Project leader & Head - (Glyco)Proteomics at Max Planck Unit for the Science of Pathogens (MPUSP)
Co-founder/CEO KPL ApS , #Proteomics researcher in Copenhagen, Denmark at Københavns Universitet, NNF Center for Protein Research #science, #technology, #culture, memes and vibes.
Associate professor at San Jose State University. PUI for life. Chaotic leader of the Wiggly Protein Lab. She/Her
Biólogo, doutor em biotecnologia (bioinformata)
Recifense morando em Belém
Aqui eu falo de ciência e reclamo do Sport
Head of Directorate at John Innes Centre. Passionate about recognising the hidden roles in research and innovation.
PI at the University of Freiburg, interested in molecular microbiology and all things related to #archaea
Structural biophysicist studying host-parasite interactions. #BeatNTD
Opinions are my own
Microbiologist @UniofYork. Former EiC of @MicrobioSoc
#MicrobioJ. Transporters, Staph, BO & skin microbiome. CSO
@morfdb.bsky.social York Knavesmire Harrier & EBOR orienteer.
@Stanford Postdoc • @UCBerkeley PhD Biophysics • @hdfcures fellow • Protein misfolding linked to neuro diseases • Single-molecule tools • Space/Pisco enthusiast