Meet the Autism Data Science Initiative grantees
The projects plan to study gene-and-environment interactions in people, stem cells and organoids, as well as predictors of positive life outcomes.
The awarded projects plan to study gene-and-environment interactions in people, stem cells and organoids, as well as predictors of positive life outcomes in autistic youth and adults.
By @callimcflurry.bsky.social
www.thetransmitter.org/spectrum/mee...
03.10.2025 21:27 — 👍 14 🔁 5 💬 0 📌 0
@biologicalpsych.bsky.social @ispg.bsky.social
03.10.2025 19:22 — 👍 0 🔁 0 💬 0 📌 0
10/ For clinicians: consider genetics as part of a precision psychiatry approach—useful for prognosis, medical surveillance, reproductive counseling, and occasionally treatment considerations tied to specific variants (see our table of variants with clinical implications).
03.10.2025 19:20 — 👍 0 🔁 0 💬 1 📌 0
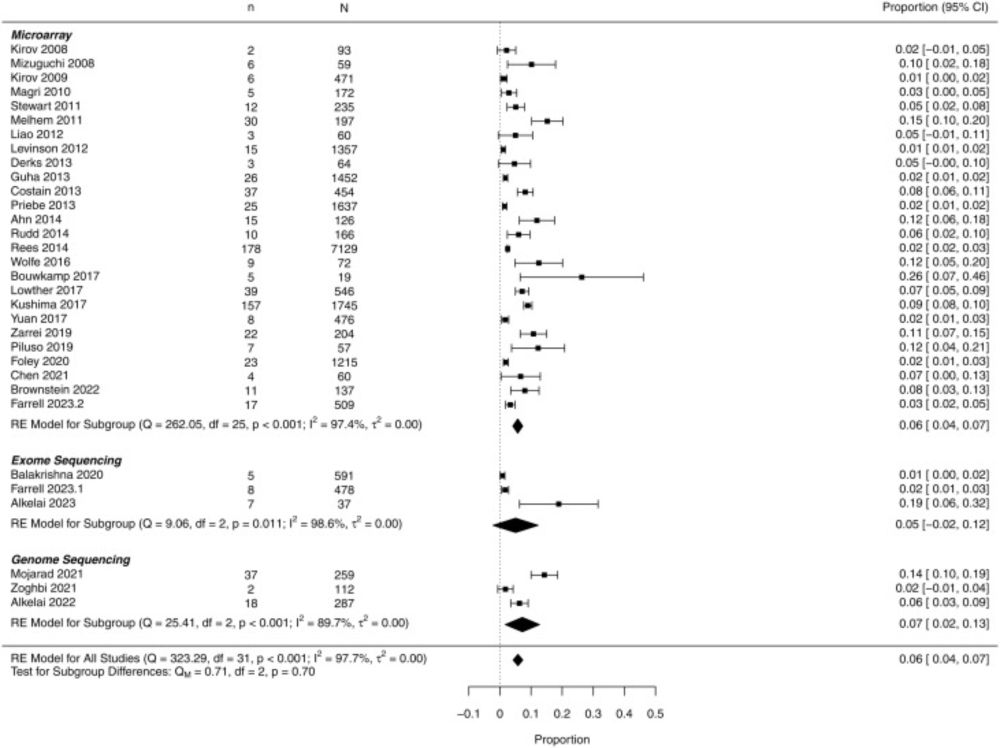
9/ What could improve yield over time: comprehensive reporting of both CNVs and SNVs, consistent ACMG/AMP interpretation, and attention to variant classes best captured by GS. As databases mature, VUS reclassification may further increase actionable returns.
03.10.2025 19:20 — 👍 0 🔁 0 💬 1 📌 0
8/ Clinical take-home: These data do not constitute a practice guideline, but they can inform diagnostic workups—especially for schizophrenia with NDD features or early onset—and motivate services to build genetics pathways and counseling capacity.
03.10.2025 19:20 — 👍 0 🔁 0 💬 1 📌 0
7/ Important caveats: substantial heterogeneity (I²≈96%), inconsistent CNV/SNV reporting across studies, and limited geographic representation (notably few data from Latin America, South Asia, Africa). The field needs better standardization and broader sampling.
03.10.2025 19:20 — 👍 0 🔁 0 💬 1 📌 0
6/ Context: The Royal College of Psychiatrists has recommended considering CMA in schizophrenia. Our pooled estimate (~6%) is higher than earlier CNV-only figures, reinforcing that genetic testing can be clinically relevant—but standards and reporting practices matter.
03.10.2025 19:20 — 👍 0 🔁 0 💬 1 📌 0
5/ Who benefits most (signal from meta-regression): higher yields in schizophrenia with co-occurring NDD features—especially intellectual disability—and earlier age of onset. These groups could be prioritized when considering clinical genetic testing.
03.10.2025 19:20 — 👍 0 🔁 0 💬 1 📌 0
4/ Key result: ~6% pooled diagnostic yield (95% CI 4–7%).
By platform: CMA ~6%, ES ~5%, GS ~7%. (Note: confidence intervals overlap; study methods & reporting varied.) This suggests ~1 in 17 patients may receive clinically informative findings.
03.10.2025 19:20 — 👍 0 🔁 0 💬 1 📌 0
3/ What we did: Systematic review & random-effects meta-analysis across MEDLINE, EMBASE, and PsycINFO (2007–2023). We pooled platform-specific yields for chromosomal microarray (CMA), exome (ES), and genome sequencing (GS), and ran meta-regressions to probe heterogeneity.
03.10.2025 19:20 — 👍 0 🔁 0 💬 1 📌 0
2/ Why this matters: genetic testing is now routine in many neurodevelopmental disorders (ID, ASD, epilepsy), yet adoption in schizophrenia has lagged—due to uncertainty about yield, variable reporting, and limited genetics training in psychiatry. We tackle that evidence gap.
03.10.2025 19:20 — 👍 0 🔁 0 💬 1 📌 0
Home | Developmental Synaptopathies Consortium
Developmental Synaptopathies Consortium works to improve the lives of patients and families affected by developmental synaptopathies.
12/ SKS & PHTS families who participated
Dr. Julian Martinez-Agosto (UCLA)
@rarediseasectn.bsky.social @rarediseasesint.bsky.social
@autismspeaks.org
@simonsfoundation.org
28.08.2025 05:32 — 👍 0 🔁 0 💬 0 📌 0
11/🏁 Takeaway:
Sensory profiles may provide a window into genetic pathogenicity across OGIDs, but variant scores alone aren’t robust prognostic tools.
Individualized neurobehavioral assessment remains essential for diagnosis, prognosis, and intervention planning.
28.08.2025 05:32 — 👍 0 🔁 0 💬 1 📌 0
10/🧩 Clinical classification:
Decision tree using behavioral + medical features (e.g., neonatal teeth for PHTS) performed above chance (CV relative error ≈0.67).
Behavioral-only tree also above chance, showing the strength of detailed phenotyping.
28.08.2025 05:32 — 👍 0 🔁 0 💬 1 📌 0
9/Combined OGIDs (SKS + PTEN + PI3K–AKT–MTOR SFARI genes):
• CADD ↗ SSP Low Energy & SSP Total
• CADD ↘ SRS-2 Total T
These were the most consistently stable correlations after 1,000 bootstrap resamples.
28.08.2025 05:32 — 👍 0 🔁 0 💬 1 📌 0
8/ PTEN-specific:
• CADD ↗ SSP Low Energy (r=0.72)
• CADD ↘ SRS-2 Total T (r=−0.64)
(both bootstrap-stable; p<0.05 uncorrected)
28.08.2025 05:32 — 👍 0 🔁 0 💬 1 📌 0
7/SKS-specific (missense only):
• REVEL ↗ SSP Auditory Filtering (r=0.77)
• AlphaMissense ↗ SSP Visual/Auditory Sensitivity (r=0.74)
• REVEL ↘ DCDQ Control During Movement (r=−0.80)
(all bootstrap-supported; p<0.05 uncorrected)
28.08.2025 05:32 — 👍 0 🔁 0 💬 1 📌 0
6/🧬 Pathogenicity scores overall:
Cross-cohort correlations were limited/inconsistent.
CADD showed the most stable associations—especially with sensory processing—supporting the need for deep phenotyping beyond variant scores alone.
28.08.2025 05:32 — 👍 0 🔁 0 💬 1 📌 0
5/🔬 Protein-domain findings:
PTEN phosphatase-domain variants → more severe social & executive deficits vs C2-domain variants.
MTOR domain differences (FAT vs PI3K) not significant (sample-size limited in SKS).
28.08.2025 05:32 — 👍 0 🔁 0 💬 1 📌 0
4/Cohorts: SKS (MTOR) n=17, PHTS (PTEN) n=74, Macrocephaly-Autism n=33, Controls n=32.
We profiled motor, adaptive, social, executive, sensory domains, ran domain-by-protein analyses, pathogenicity–phenotype correlations, and built diagnostic decision trees.
28.08.2025 05:32 — 👍 0 🔁 0 💬 1 📌 0
3/Our central question: How much clinical and genotype–phenotype overlap exists across disorders in the same pathway? Given their rarity, can analyzing them together reveal new insights to improve diagnosis & care?
28.08.2025 05:32 — 👍 1 🔁 0 💬 1 📌 0
2/Smith-Kingsmore Syndrome (SKS) is caused by MTOR variants; PTEN Hamartoma Tumor Syndrome (PHTS) by PTEN variants.
Both are overgrowth–intellectual disability syndromes (OGIDs) in the PI3K–AKT–MTOR pathway.
28.08.2025 05:32 — 👍 0 🔁 0 💬 1 📌 0
Structural variants are significant contributor to autism. But many SVs & TRs are hard to detect with short reads. Long read sequencing with @pacbio.bsky.social and @nanoporetech.com captures and maps out alot of what short reads miss. So what can LR-WGS tell us about autism? 🧵
23.07.2025 22:53 — 👍 22 🔁 6 💬 3 📌 2
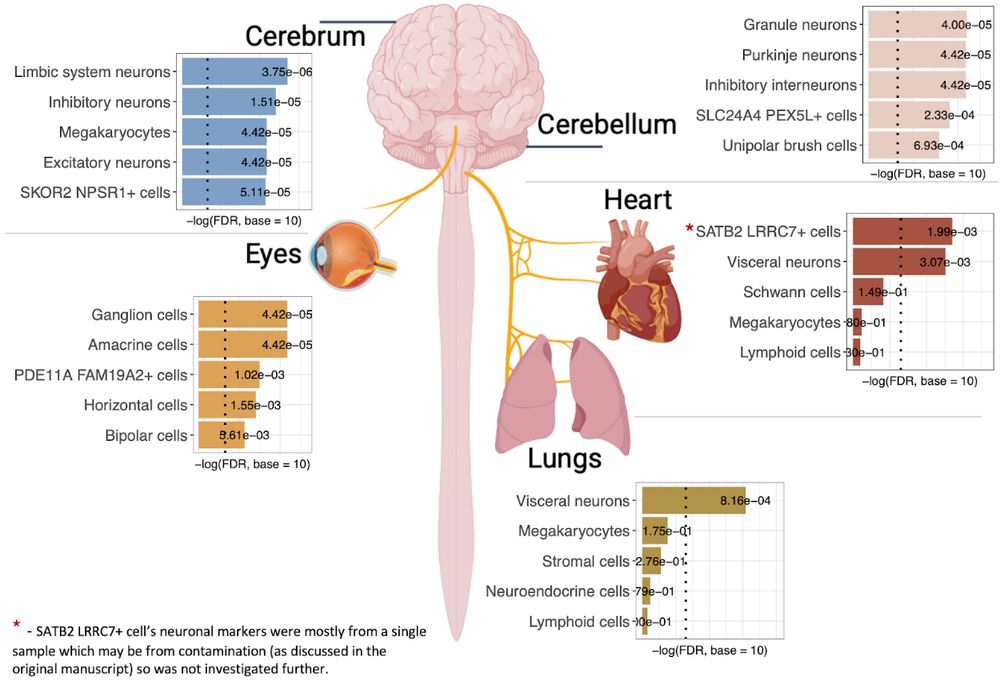
Excited about our new preprint: 1st successful genome-wide study of >61,000 panic attack and 29,000 panic disorder cases. www.medrxiv.org/content/10.1... We find 17 associations & evidence that peripheral neurons in eye, lungs and heart are involved in panic & other psychiatric disorders 1/n
17.06.2025 10:50 — 👍 47 🔁 24 💬 1 📌 4
Official account for the Biological Psychiatry family of journals.
Publishing cutting-edge research in psychiatric neuroscience, cognitive neuroimaging, and global open science.
Research, news, and commentary from Nature, the international science journal. For daily science news, get Nature Briefing: https://go.nature.com/get-Nature-Briefing
PhD, MD, psychiatry resident.
She/her.
Associate Professor @mcgill.ca
Study neurodevelopmental disorders using mouse and human organoid models
https://www.huanglabmcgill.org/
Based at the University of Queensland, our research aims to improve accessibility and equity of genomic testing across all healthcare settings🧬
Join the fight to cure #mentalillness. Since 1987 BBRF has awarded more than $475 million in #mentalhealth research grants worldwide. Give the gift of recovery: https://bbrfoundation.org/donate?BlueSky
Head of Psychosis Research Unit - Translational research in Child & Adolescent psychiatry - Lausanne University Hospital
Director of bioinformatics at AstraZeneca. subscribe to my youtube channel @chatomics. On my way to helping 1 million people learn bioinformatics. Educator, Biotech, single cell. Also talks about leadership.
tommytang.bio.link
http://www.lifespanlab.com/
Follow us to stay up to date with the American Journal of Medical Genetics Part B (Neuropsychiatric Genetics). @ispg.bsky.social Affiliate Journal.
Edited by: Stephen J. Glatt
shendure lab |. krishna.gs.washington.edu
Your friendly neighborhood neuroinformatics scientist at the NIMH Data Science & Sharing Team and Dad/Husband at Home, opinions are my own
Statistical geneticist. Professor of Human Genetics and Biostatistics at the University of Pittsburgh. Assiduously meticulous.
Minds, moods, and molecules ␥ Psychological and psychiatric complex trait genetics ␥ Researcher in Edinburgh, Scotland
https://differentialist.info/about/
PhD student in statistical genetics at Vrije Universiteit Amsterdam
asst prof | clinical psychologist | some sort of geneticist | not a neuroscientist | mass general & harvard med | engagement ≠ endorsement
Human Genetics, Molecular Epidemiology, Biological Psychiatry
https://medicine.yale.edu/lab/polimanti/
Molecular Psychiatry, Neuroscience, Genetics, Professor at Karolinska Insitutet
Associate Professor, University of Utah
Statistical genetics, psychiatric genetics, bioethics
Chair, PGC Suicide Working Group