Congratulations to Alok and all the authors!
@broadinstitute.org @mgbresearch.bsky.social
22.01.2026 20:26 — 👍 0 🔁 0 💬 0 📌 0

@broadinstitute.org
@mgbresearch.bsky.social
16.01.2026 20:06 — 👍 0 🔁 0 💬 0 📌 0

These studies firmly establish a foundation for development of an inhibitor of CARD9 inflammatory response. More broadly, this approach illustrates a strategy for using human genetics and chemical biology to guide small-molecule discovery against challenging immune targets. (7/7)
16.01.2026 20:06 — 👍 1 🔁 0 💬 1 📌 0


The optimized compound blocked CARD9-dependent NFκB activation and cytokine release in human dendritic cells and reduced inflammation in mouse model of fungal infection expressing humanized CARD9 (hCARD9), demonstrating the feasibility of pharmacological targeting of CARD9. (6/7)
16.01.2026 20:06 — 👍 1 🔁 0 💬 1 📌 0


Using time-resolved FRET (TR-FRET) displacement screens, surface plasmon resonance (SPR), and structure-activity relationship studies we identified a benzodiazepine compound with selective CARD9 binding and cellular activity. (5/7)
16.01.2026 20:06 — 👍 1 🔁 0 💬 1 📌 0


To explore pharmacological modulation of CARD9, we took a binder-first strategy: a DNA-encoded library screen and subsequent structural studies revealed a ligandable pocket in the CARD9 coiled-coil domain, guiding subsequent chemical exploration. (4/7)
16.01.2026 20:06 — 👍 1 🔁 0 💬 1 📌 0

In prior work, we leveraged CARD9Δ11 to study CARD9 regulation and identified the E3 ligase TRIM62 as a CARD9-interacting protein that regulates CARD9 activation through post-translational modification. (3/7)
www.sciencedirect.com/science/arti...
16.01.2026 20:06 — 👍 1 🔁 0 💬 1 📌 0
CARD9 is a key immune adaptor required for immune responses against fungi and bacteria and is implicated in multiple inflammatory diseases. The truncation variant CARD9Δ11 protects against Crohn’s disease, suggesting CARD9 inhibition as a strategy for therapeutic intervention. (2/7)
16.01.2026 20:06 — 👍 1 🔁 0 💬 1 📌 0
Fun collaboration with @thexavierlab.bsky.social and Ribbeck groups to figure out how this human lectin plays multiple roles in controlling the #microbiome: stabilizing the mucus barrier and reducing bacteria that breach it. news.mit.edu/2026/protein... #glycotime
15.01.2026 15:26 — 👍 14 🔁 6 💬 0 📌 0
@mgbresearch.bsky.social
07.01.2026 20:58 — 👍 0 🔁 0 💬 0 📌 0
Very excited to share our latest work in @Nature integrating single-cell & spatial transcriptomics with bidirectional CRISPR screens to unravel an immune–stromal cell circuit dependent on the transcription factor GLIS3 that regulates intestinal fibrosis during chronic inflammation. Congrats, Vlad!
07.01.2026 20:58 — 👍 5 🔁 2 💬 1 📌 0

IHMC 2026 SEOUL
IHMC 2026 SEOUL 사이트입니다.
Register now for the International Human Microbiome Consortium (#IHMC) 2026, June 3–5 in Seoul, Korea!
Join this global gathering of researchers for an exciting program highlighting the latest discoveries in human #microbiome science:
www.ihmc2026seoul.com/html/
11.12.2025 18:34 — 👍 0 🔁 0 💬 0 📌 0
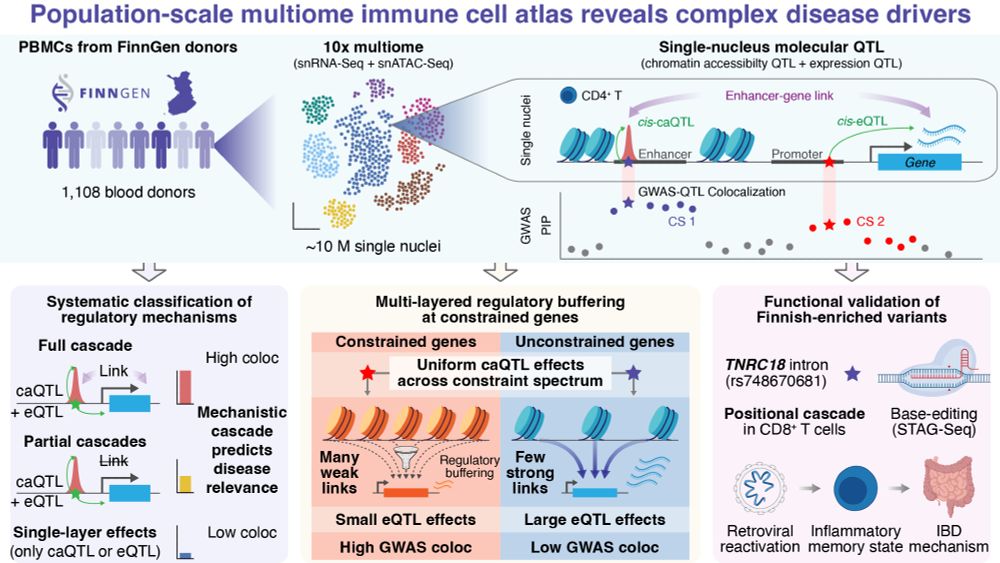
Excited to share our new FinnGen single-nucleus multiome preprint! 🧬
We profiled ~10M PBMCs (snRNA-seq + snATAC-seq) from 1,108 Finnish donors to map how genetic variants drive complex disease through chromatin and gene regulation 🧵👇
🔗 Link: www.medrxiv.org/content/10.1...
01.12.2025 15:36 — 👍 33 🔁 19 💬 1 📌 0

ICMI 2026
ICMI provides an ideal opportunity to showcase your organization’s products and services and have direct interface with mucosal immunologists from around the globe.
Registration now open for the International Congress of Mucosal Immunology (ICMI) 2026, Jul 13–17 in Montreal, Canada.
www.socmucimm.org/meetings-eve...
Looking forward to being part of this lineup of talks highlighting research at the interface of the immune system and mucosal surfaces!
04.12.2025 15:04 — 👍 2 🔁 0 💬 0 📌 0
Protein language models uncover carbohydrate-active enzyme function in metagenomics
Check out their findings from paired mother/infant microbiome & disease-specific metagenomic datasets! rdcu.be/eRVYz (2/2)
01.12.2025 19:15 — 👍 0 🔁 0 💬 0 📌 0
Protein language models uncover carbohydrate-active enzyme function in metagenomics
Kumar Thurimella & co-authors addressed the challenge of functionally annotating uncharacterized microbial enzymes with CAZyLingua, a deep learning model for classifying CAZymes from metagenomics. (1/2)
01.12.2025 19:15 — 👍 1 🔁 0 💬 1 📌 0
JCI -
Allosteric modulation of the solute carrier transporter SLC39A8 potentiates manganese and cadmium uptake
Solute carrier (SLC) transporters are implicated in many human diseases. In @jclinicalinvest, we show that efavirenz potentiates metal transporter SLC39A8 via a cryptic allosteric pocket—revealing new potential for therapeutic targeting.
www.jci.org/articles/vie...
04.11.2025 20:52 — 👍 2 🔁 1 💬 0 📌 0

In Nov issue: rupress.org/jem/issue/22...
@thexavierlab.bsky.social describe the biological function of Ankrd55 in mice, working as a functional modulator of T cell metabolism impacting Th17 responses. Cover shows a colon section of a mouse that received Ankrd55−/− T cells doi.org/10.1084/jem....
03.11.2025 15:04 — 👍 2 🔁 1 💬 0 📌 0

Congrats to Jinjin Xu & co-authors – an image from their recent @jem.org article that describes Ankrd55 as a modulator of T cell metabolism impacting Th17 responses was selected for the cover!
Read the study here: rupress.org/jem/article-...
@broadinstitute.org @mgbresearch.bsky.social
04.11.2025 14:47 — 👍 2 🔁 2 💬 1 📌 0
Congrats to the authors!
@broadinstitute.org
@mgbresearch.bsky.social
30.10.2025 16:18 — 👍 0 🔁 0 💬 0 📌 0

🔦 Spotlight! The Korean Association of Immuologists International 2025 is convening Oct 30th-Nov 1st in Incheon, South Korea. KAI 2025 will address current topics in immunology, encompassing both fundamental research and its application to immunological diseases.
kai2025.kr
28.10.2025 20:33 — 👍 0 🔁 0 💬 0 📌 0

Harvard Medical School 2025 State of the School Address
In the 2025 State of the School Address, Dean George Q. Daley described the challenges and opportunities ahead for Harvard Medical School and academic research.
In his “Harvard Medical School 2025 State of the School Address”, Dean George Daley highlighted our collaboration with the Clardy lab studying the connection between the gut microbiome and depression @harvardmed.bsky.social @broadinstitute.org
#vimeo #microbiome #depression #gut
25.09.2025 18:31 — 👍 0 🔁 0 💬 1 📌 0
Chemical biologist at MIT, passionate about glycoscience and all chemical biology
The largest hospital-based #research network in the U.S.
Where #science and #medicine converge to improve patient care.
Blog: https://mgriblog.org
We ❤️🔬
Neuroscientist passionate about Glia and Neuroimmunology & immune-brain cross talk in health & disease.
Foodie & Wine enthusiast. #openscience and collaboration
Instructor in Medicine, MGH / HMS / Broad Institute
https://mkanai.github.io/
Working to realize the promise of human genomics to better predict, prevent, and treat cancer. For more info: https://labs.dana-farber.org/collins-genomics/
Professor of Microbiology @ UMass Chan Medical School | Host-Microbe Interactions | Bacteria | Immunology | Artificial Intelligence
Immunology Ph.D. student focusing on neuro-immune interactions in the gut.
Anti-apartheid.
Views are my own.
Postdoc | Broad Institute | Gut microbiome
• 🇧🇷 Assist Prof at MGH / Harvard Medical School 🇺🇸
• Postdoc from EPFL with Alex Persat🇨🇭
• PhD from Caltech with Dianne Newman 🇺🇸
• 🧫 Microbial physiology | Host-pathogen interactions | AMR | Tissue engineering | Organoids 🫁 (he/him)
Journal of Experimental Medicine publishes immunology, cancer, stem cells, microbial pathogenesis, vascular biology, and neurobiology research. Published by Rockefeller University Press @rupress.org
🌐 rupress.org/jem
A forerunner in the global landscape of genomics research projects. The study has collected and analysed genome and health data from >500,000 Finnish biobank donors to understand the genetic basis of diseases.
www.finngen.fi
Physician-Scientist at Massachusetts General Hospital. Posting about biology (mostly immunology) and medicine
https://www.rahimi-lab.com
A multidisciplinary community of researchers with the mission to better understand the roots of disease and narrow the gap between new biological insights and impact for patients. Broadinstitute.org
Immunology-focused research in cancer & aging | understanding immune evasion & resilience | improving health outcomes. 🔗 https://genelayinstitute.org/
#Brigham&WomensHospital #MassGeneralHospital @HarvardMed.bsky.social
Lab of Vijay Kuchroo at @GeneLayInstitute.bsky.social @MGBResearch.bsky.social @HarvardMed.bsky.social @BroadInstitute.org
🧪 #Immunosky
🔗 kuchroolab.bwh.harvard.edu
Posts by lab members unless signed by VKK. Views are our own.
Immunologist at Benaroya Research Institute, Seattle WA
https://www.harrison-lab.org
Associate Professor at Harvard Medical School, in love with a red-haired beautiful woman, his kids... and immunology. He/him/his. 🇮🇹 🇺🇸 Views my own.
Senior Scientific Editor at Immunity, Cell Press @cp-immunity.bsky.social; fan of science (especially immunology), space, & nature; views are my own
https://www.cell.com/immunity/home
Scientist, Dog mom, Sushi Queen, and New Yorker
Naiklab.com
Views are my own





















