Our story on mechanosensation in baroreceptor cells of the kidney is finally out! Well worth the wait on this project we began in 2020 that turned into a massive collaborative effort within the kidney field. 🫘 www.sciencedirect.com:5037/science/arti...
04.12.2025 16:42 — 👍 45 🔁 17 💬 1 📌 0
It was great mentoring and working with Venkat Sankar on this project. Also great working with Howard Chang, Paul Mischel, @agnanam.bsky.social, Ivy Wong, and other collaborators on this project. 11/11
19.11.2025 19:03 — 👍 3 🔁 0 💬 0 📌 0

Overall, we propose that ecDNA is not only selected in cancer because of oncogenes, it is also actively retained because of retention elements, allowing it to persist in a growing cancer cell population. 10/11
19.11.2025 19:03 — 👍 3 🔁 0 💬 1 📌 0

Finally, retention elements are unmethylated on ecDNA and when we methylate them, ecDNA becomes untethered from chromosomes. Plasmids with methylated retention elements are also no longer retained. 9/11
19.11.2025 19:03 — 👍 2 🔁 0 💬 1 📌 0
We found that most ecDNAs in patient tumors contain retention elements. ecDNA size is much bigger than the oncogenes themselves but is anti-correlated with abundance of retention elements in the surrounding locus, suggesting selection of these elements in the retained ecDNA. 8/11
19.11.2025 19:03 — 👍 1 🔁 0 💬 1 📌 0
Retention elements on ecDNA interact with previously identified mitotic bookmarks on chromosomes in chromatin conformation analysis, suggesting that retention elements may interact with chromosomes in mitosis via active DNA sites, potentially mediated by bookmarking factors. 7/11
19.11.2025 19:03 — 👍 1 🔁 0 💬 1 📌 0


Using live cell imaging, we found that adding a retention element to a plasmid makes it more likely to attach to chromosomes during mitosis. 6/11
19.11.2025 19:03 — 👍 2 🔁 0 💬 1 📌 0

We verified that these elements, when individually cloned into a plasmid, can promote retention of the plasmid in cells. Putting more copies of an element in a plasmid increases its retention additively. 5/11
19.11.2025 19:03 — 👍 2 🔁 0 💬 1 📌 0
We found that many of the retained DNA elements, which we call “retention elements”, are active regulatory sequences such as promoters and enhancers. This was exciting to us because these are genomic sites where many DNA-binding proteins can interact with DNA. 4/11
19.11.2025 19:03 — 👍 1 🔁 0 💬 1 📌 0

To identify elements within ecDNA that may act as anchors for hitchhiking, we shattered the entire human genome into short pieces of DNA, cloned them into a plasmid pool and asked which DNA pieces can be retained over many cell passages. 3/11
19.11.2025 19:03 — 👍 2 🔁 0 💬 1 📌 0

ecDNA does not have centromeres but is inherited along with chromosomes by tethering to them. Although this “hitchhiking” phenomenon has been observed for many years, the mechanism is unknown. 2/11
19.11.2025 19:03 — 👍 1 🔁 0 💬 1 📌 0
🎉 Congratulations to @kinglhung.bsky.social from team eDyNAmiC for receiving the prestigious prize and for having his essay, “Oncogenes out of context: Cancer genes break free from the regulatory constraints of chromosomes”, published in
@science.org.
📖 Read more - lnkd.in/ekMqXC6J
18.11.2025 16:09 — 👍 2 🔁 1 💬 0 📌 0

Oncogenes out of context: Cancer genes break free from the regulatory constraints of chromosomes
Cancer genes break free from the regulatory constraints of chromosomes
Congrats King Hung, Science & SciLifeLabs award winner @stanfordmedicine.bsky.social, collaborating with @stanford-chemh.bsky.social institute scholar @paulmischel.bsky.social
www.science.org/doi/10.1126/...
14.11.2025 17:49 — 👍 17 🔁 5 💬 0 📌 0

Honored to receive the Science & SciLifeLab Prize! Very grateful to all my mentors and colleagues. I wrote an article in @science.org about my PhD thesis research on extrachromosomal DNA at Stanford @stanfordmedicine.bsky.social. www.science.org/doi/10.1126/...
14.11.2025 17:08 — 👍 35 🔁 4 💬 1 📌 1
And here is the story after peer review. This work highlights our current approach, which is to use Piezo channels to uncover new areas of biology that are shaped by mechanical forces: www.science.org/doi/10.1126/...
13.11.2025 22:05 — 👍 77 🔁 29 💬 2 📌 4

Dear Fly Community,
In May 2025, the NIH terminated all grant funding to Harvard University, including the NHGRI grant that supported FlyBase. This grant also funded FlyBase teams at Indiana University (IU) and the University of Cambridge (UK), and as a result, their subawards were also canceled.
The Cambridge team has secured support for one to two years through generous donations from the European fly community, emergency funding from the Wellcome Trust, and support from the University of Cambridge. At IU, funding has been secured for one year thanks to reserve funds from Thom Kaufman and a supplement from ORIP/NIH to the Bloomington Drosophila Stock Center (BDSC).
Unfortunately, the situation at Harvard is far more critical. Harvard University had supported FlyBase staff since May but recently denied a request for extended bridge funding. As a result, all eight employees (four full-time and four part-time) were abruptly laid off, with termination dates ranging from August to mid-October depending on their positions. In addition, our curator at the University of New Mexico will leave her position at the end of August. This decision came as a shock, and we are urgently pursuing all possible funding options.
To put the need into perspective: although FlyBase is free to use, it is not free to make. It takes large teams of people and millions of dollars a year to create FlyBase to support fly research (the last NHGRI grant supported us with more than 2 million USD per annum).
To help sustain FlyBase operations, we have been reaching out to you to ask for your support. We have set up a donation site in Cambridge, UK, to which European labs have and can continue to contribute, and a new donation site at IU to which labs in the US and the rest of the world can contribute. We urge researchers to work with their grant administrators to contribute to FlyBase via these sites if at all possible, as more of the money will go to FlyBase. However, we appreciate that some fu…

https://wiki.flybase.org/wiki/FlyBase:Contribute_to_FlyBase
Our immediate goals are:
1. To maintain core curation activities and keep the FlyBase website online
2. To complete integration with the Alliance of Genome Resources (The Alliance).
Integration with the Alliance is essential for FlyBase’s long-term sustainability. For nearly a decade, NHGRI/NIH has supported the unification of Model Organism Databases (MODs) into the Alliance, which we aim to achieve by 2028. Therefore, securing bridge funding to sustain FlyBase over the next three years is crucial for successful integration and the long-term access to FlyBase data.
At present, our remaining funds will allow us to keep the FlyBase website online for approximately one more year. Beyond that, its future is uncertain unless new funding is secured. We will, of course, continue pursuing additional grant opportunities as they arise.
Given the uncertainty of future NIH or alternative funding sources, we are relying on the Fly community for support. Your contributions will directly help us retain the staff needed to complete this transition and to secure ongoing fly data curation into the Alliance beyond 2028.
We at FlyBase are incredibly grateful for the outpouring of support from the community during this challenging time. Your encouragement has strengthened our resolve and underscores how vital this resource remains to Drosophila research worldwide.
Sincerely,
The FlyBase Team
The community of Drosophila researchers is amazing, mutually supportive and collaborative. Right now a key resource for our community, @flybase.bsky.social , is threatened by the cancellation of its NIH grant and is seeking community help in raising short term funds 1/n 🧪 please share
23.08.2025 12:18 — 👍 150 🔁 127 💬 1 📌 6

Cool approach to isolate extrachromosomal DNA: karyotyping by FACS! Wish I had thought of it. www.biorxiv.org/content/10.1...
28.03.2025 04:28 — 👍 2 🔁 0 💬 0 📌 0
So excited for you, Rose!
27.03.2025 16:15 — 👍 2 🔁 0 💬 0 📌 0
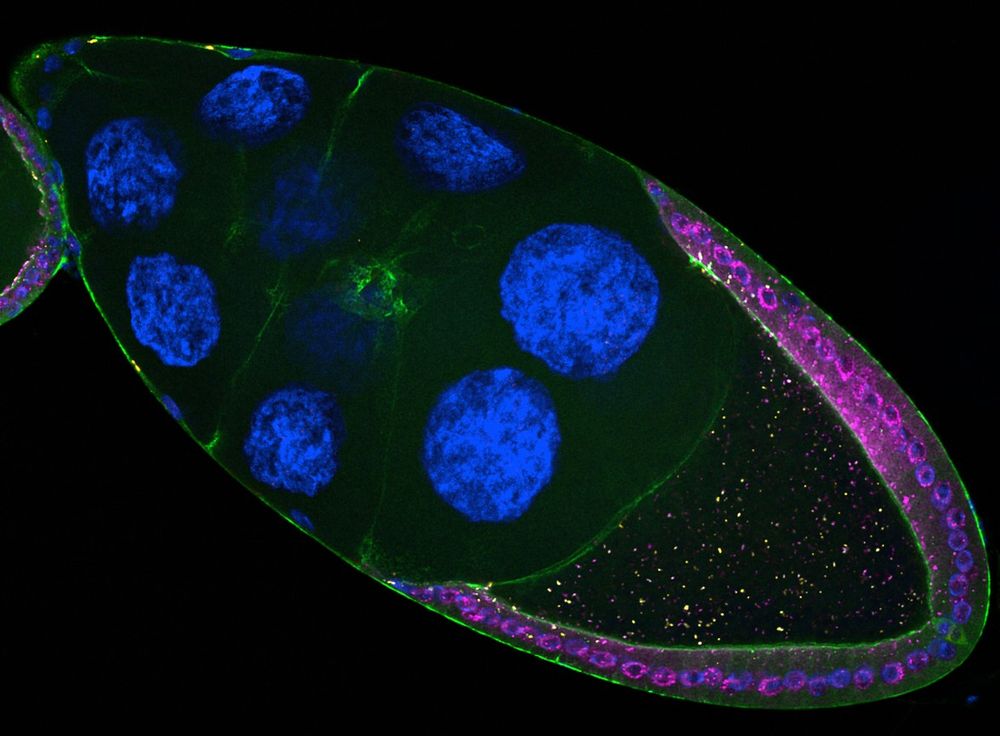
Drosophila follicle showing retrotransposons (pink & yellow) expressed in somatic cells infecting the oocyte
1/ Transposable elements are often called "jumping genes" because they mobilize within genomes. 🧬
But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
17.03.2025 11:56 — 👍 546 🔁 259 💬 12 📌 33
if you've had an NIH grant terminated, I want to hear about it. Signal: katherinejwu.12
07.03.2025 14:17 — 👍 493 🔁 384 💬 3 📌 7

a cat is laying on top of the number 25
ALT: a cat is laying on top of the number 25
The Patapoutian lab started 25 years ago on March 1, 2000. A quarter-century of science with brilliant trainees, colleagues & collaborators. Cheers to my mentors & mentees! Excited to celebrate with past & present lab members in November. Here’s to more great science 🧪 and better times ahead!
01.03.2025 15:14 — 👍 168 🔁 7 💬 10 📌 2
Flooding BlueSky with good news today. Neal lab received a NoA! Grateful our NIH R35 MIRA renewal is funded to support our ongoing work on membrane protein quality! 🙏🙏
14.02.2025 23:28 — 👍 552 🔁 38 💬 27 📌 3
Opportunity to work with an amazing scientist!
10.01.2025 01:52 — 👍 2 🔁 0 💬 0 📌 0
Join our lab! We're hiring a research technician—a great opportunity for individuals with a Bachelor's degree who want to gain valuable research experience before heading to medical or graduate school.
recruiting2.ultipro.com/SCR1003TSRI/...
16.12.2024 23:14 — 👍 30 🔁 19 💬 1 📌 2
Thanks for highlighting our work!
19.11.2024 21:58 — 👍 0 🔁 0 💬 0 📌 0
Also recently published work from this month:
bsky.app/profile/king...
19.11.2024 17:14 — 👍 1 🔁 0 💬 0 📌 0
Hi bluesky! A re-introduction: I am a cell biologist interested in cellular signaling logic and decision making. I worked on cancer genomes in my PhD at Stanford, and am now a postdoc working on how cells interpret force at Scripps in the Patapoutian lab. Excited to reconnect here!
19.11.2024 17:14 — 👍 15 🔁 1 💬 1 📌 0
Group leader @mpi-mg, Berlin
Chromatin tracing, epigenetics, development. Equity and advocacy in academia.
Data Scientist @ Stanford Health Care and perpetual MD/PhD student. 🏳️🌈
AI/ML for health, #rstats and #pydata
Postdoc at Scripps Research. Incoming assistant professor at Northwestern University. Investigating body-brain communication.
Neuroscientist studying mechanical interoception. Views are my own.
Prof @ Harvard University, Broad Institute, and Wyss Institute.
Synthetic & systems biology.
Trying to understand the living world by engineering a synthetic one.
Lab website: bu.edu/khalillab
MD/PhD Student at Stanford interested in immune signaling, protein engineering, autoimmunity and cancer. PhD with Chris Garcia on a Hertz fellowship.
all things cytoskeleton - nuclear actin dynamics and genome - nucleoskeleton and nuclear organisation - cancer cell invasion and pharmacology
@University of Freiburg
Studying cells inside embryos since way before it was cool.
https://www.wallingfordlab.org/
Asst Prof | Harvard
Freeman Hrabowski Scholar | HHMI
Founder | Leading Edge @leadingedgeprogram.bsky.social
My lab studies menstruation and menstrual disorders.
www.mckinleylab.org
Opinions are my own and do not reflect those of my employer.
Neuroscience, meninges, blood-brain barrier, cake (or cookies)
http://siegenthalerlabcu.weebly.com/
Award-winning home tank for all things zebrafish! Learn more about us and our Slack tankspace at our website: linktr.ee/zebrafishrock
Dad, bioengineer, climber, skier, surfer (in that order, more or less).
Chaotic good alignment, or so my lab tells me...
Genomics, Machine Learning, Statistics, Big Data and Football (Soccer, GGMU)
Professor at the NYU School of Medicine (https://yanailab.org/). Co-founder and Director of the Night Science Institute (https://night-science.org/). Co-host of the 'Night Science Podcast' https://podcasts.apple.com/us/podcast/night-science/id1563415749
Associate Prof @HarvardMed. Microbial evolution, antibiotic resistance, mobile genetic elements, algorithms, phages, molecular biotech, etc. Basic research is the engine of progress.
baymlab.hms.harvard.edu
Science integrity consultant and crowdfunded volunteer, PhD.
Ex-Stanford University. Maddox Prize/Einstein F Award winner
NL/USA/SFO.
#ImageForensics
@MicrobiomDigest on X.
Blog: ScienceIntegrityDigest.com
Support me: https://www.patreon.com/elisabethbik
Postdoc @RouxLab.bsky.social investigating topological defects and morphogenesis. PhD was on tensile stresses and endothelial cells.
@dfg.de Walter Benjamin Fellow & Postdoctoral Researcher @yap-lab.bsky.social | Co-organizer @epimechfc.bsky.social | Mechanobiology - Cell & Tissue Mechanics - Soft Matter Physics - Image Analysis
👉 https://julia-eckert.github.io
@AIOpticalBioLab.bsky.social. Group Leader #ITQB & H. Prof #UCL. #Nanoscopy & #AI for #LifeSciences. #SRRF sensei. #ERCCoG, #EMBO member. Dad. Opinions my own
- henriqueslab.org -
Chief Editor of Nature Biomedical Engineering. Ph.D. Previously at Nature Methods. @rita_strack on the place formerly known as twitter.
















