
(10/N) If you are interested check out our preprint and code! Thanks for reading!
Arxiv: arxiv.org/abs/2502.07746
Code: github.com/Krishnaswamy...
@krishnaswamylab.bsky.social
We develop AI methods for science, particularly deep learning methods based on data geometry, topology and dynamics systems.

(10/N) If you are interested check out our preprint and code! Thanks for reading!
Arxiv: arxiv.org/abs/2502.07746
Code: github.com/Krishnaswamy...
(9/N) Grateful to lead authors Siddharth Viswanath and Hiren Madhu, and coauthors Dhananjay Bhaskar, Jake Kovalic, Dave Johnson, Chris Tape, Ian Adelstein, Rex Ying and Michael Perlmutter!
07.11.2025 14:19 — 👍 0 🔁 0 💬 1 📌 0(8/N) ✨ In short, HiPoNet is a neural network that takes in a whole point cloud, and utilizes methods from geometry and topology to derive features of the point cloud.
→ Multiple learned views
→ Simplicial wavelets to extract multi-scale structure

(7/N) What’s more, HiPoNet’s learned feature weights are interpretable. For example, immune-related markers like CD11b, CD118, and FOXP3 consistently emerge as important across views, aligning with known tumor-immune biology.
07.11.2025 14:17 — 👍 0 🔁 0 💬 1 📌 0
(6/N) We find that HiPONet consistently outperforms other methods on these datasets.
07.11.2025 14:16 — 👍 0 🔁 0 💬 1 📌 0
(5/N) We then evaluate HiPOnet on patient level classification tasks using single cell and spatial proteomics data:
07.11.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0
(4/N) We first check that HiPoNet preserves topological features of datasets by predicting persistence from the datasets:
07.11.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0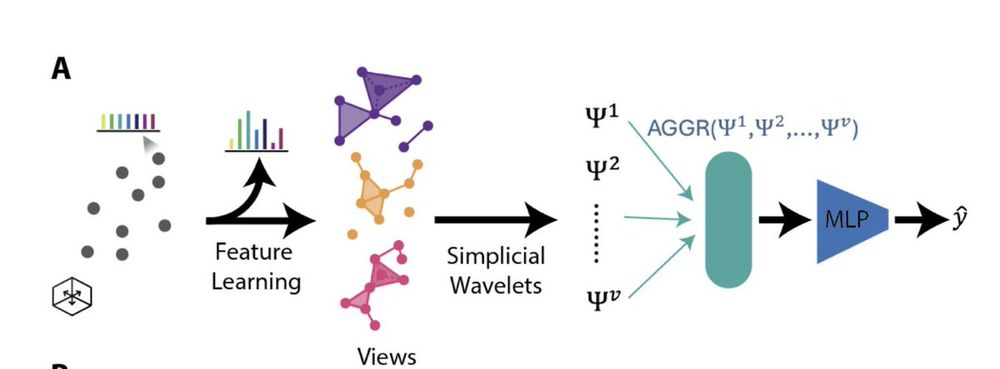
(3/N) HiPoNet models each cloud as a simplicial complex, and uses simplicial wavelet transforms to capture point-cloud level embeddings. Not only that---it uses multiview learning, i.e. different projections of the features to capture different underlying factors in the data.
07.11.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0(2/N) Modern technologies such as mass cytometry or scRNA-seq now allow large cohorts of patients to be measured. Each patient is actually a large single cell dataset creating a high-dimensional point cloud. But these point clouds are far more complex than 3D shapes that ML methods have handled..
07.11.2025 14:13 — 👍 0 🔁 0 💬 1 📌 0
(1/N) Thrilled to share that our paper HiPoNet (High dimensional Point cloud Network) to be presented at NeurIPS 2025! HiPoNet treats an entire high-dimensional point cloud as a datapoint! It captures multi-scale geometry and topology of the cloud perform classification and regression tasks.
07.11.2025 14:09 — 👍 11 🔁 3 💬 1 📌 0Good morning from Vancouver! 🌞⛰️ We’re at #ICML2025 presenting at GenBio + MAS. Come chat with us about machine learning in biology!
With @RobertXiangruTang, @ChenLiu, @DanqiLiao. Views like this make the science even sweeter.🧬🧠
#ML #ComputationalBiology #ICML #Vancouver
Defining and benchmarking open problems in single-cell analysis - @krishnaswamylab.bsky.social @fabiantheis.bsky.social bit.ly/4l8dsiF
01.07.2025 21:01 — 👍 33 🔁 9 💬 1 📌 0
Flyer for the Kavli Awards Research in Progress of June 23 showing pictures of the speakers in the Kavli logo, with a schedule: 12:00 pm Dhananjay Bhaskar, PhD 2024 Kavli Postdoctoral Fellowship Krishnaswamy & Colón-Ramos labs 12:20 pm LaShae Nicholson, PhD 2022 Kavli Award for Academic Diversity Strittmatter lab 12:40 pm José Jaime Martínez-Magaña, PhD 2023 Kavli Award for Academic Diversity Montalvo-Ortiz lab 1:00 pm Kevin Chen, PhD 2024 Kavli Postdoctoral Fellowship Emonet & Clark labs 1:20 pm break 1:30 pm Aaron Kuan, PhD & Joerg Bewersdorf, PhD 2024 Kavli Teams Award 1:50 pm Enock Teefe, MD 2023 Kavli Award for Academic Diversity Pittenger & Fernandez labs 2:10 pm Hongyan Hao, PhD 2024 Kavli Postdoctoral Fellowship Hammarlund & De Camilli labs
Mark your calendars 🗓️ for June 23 for a Kavli Awards Research in Progress!🧠
Hear from Drs. Bhaskar (@krishnaswamylab.bsky.social), @lashaeneuroxp.bsky.social, Chen (@emonetlab.bsky.social), Martinez-Magaña (@janitzamontalvo.bsky.social), @kuanawanda.bsky.social, Teefe, & @haohy.bsky.social.

New preprint! Excited to share our latest work “Accelerated learning of a noninvasive human brain-computer interface via manifold geometry” ft. outstanding former undergraduate Chandra Fincke, @glajoie.bsky.social, @krishnaswamylab.bsky.social, and @wutsaiyale.bsky.social's Nick Turk-Browne 1/8
03.04.2025 23:04 — 👍 66 🔁 20 💬 2 📌 3Excited to organize this! Should be an exciting event!!
26.01.2025 13:55 — 👍 11 🔁 0 💬 0 📌 0Great ideas to introduce to the community!! Enjoyed the talk!
25.01.2025 00:40 — 👍 4 🔁 0 💬 0 📌 0
Join us for an exciting session in @jointmath.bsky.social tomorrow! Here is our revised schedule!
10.01.2025 19:19 — 👍 15 🔁 1 💬 0 📌 0We are at @jointmath.bsky.social Jan 8-11!
1/8: My talk at Topological Machine Learning I
1/8: Dhananjay Bhaskar at Contributed Paper session on Topology
1/11 join our special session on Mathematical and Computational Oncology
Also check out @pseudomanifold.topology.rocks on 1/9!!



Merry Xmas!
25.12.2024 15:15 — 👍 6 🔁 0 💬 0 📌 0
Looking for talented postdocs in deep learning for neuroscience and neuromodulation with the Murat Gunel lab in neurosurgery (medicine.yale.edu/lab/gunel/)! Projects include brain signal decoding, dynamics modeling for neuromodulation ASD and other conditions. See: krishnaswamylab.org/join
24.12.2024 15:05 — 👍 9 🔁 2 💬 0 📌 1Frohe Weinachten!
24.12.2024 15:00 — 👍 1 🔁 0 💬 0 📌 0
(10/n) For code see github.com/Krishnaswamy.... Thanks for reading!
21.12.2024 17:57 — 👍 1 🔁 0 💬 0 📌 0
(9/n) GSPA-Pt can be used to map patient sample manifolds. We mapped 48 melanoma patient scRNA-seq samples and classified response from the patient embedding using logistic regression. The GSPA-Pt gene embeddings achieved the highest classification performance.
21.12.2024 17:57 — 👍 3 🔁 0 💬 1 📌 0
(8/n) GSPA-LR concatenates ligand (L) and receptor (R) for a pair representation. LR pair modules captures a diverse range of LR profiles, finer the cluster-level analysis. For example, in skin cells, Module 5 includes Ccl5–Ccr5 link present in AG and AG CPI, epithelial, myeloid and T cells.
21.12.2024 17:55 — 👍 1 🔁 0 💬 1 📌 0
(7/n) With the Nik Joshi Lab at Yale we presented a new dataset of 39K CD8+ cells from LCMV infections. Interestingly only GSPA-based gene localization finds genes associated with type 1 interferon signaling. DE requires clustering, but DE genes in clusters do not reveal this signature!
21.12.2024 17:53 — 👍 1 🔁 0 💬 1 📌 0
(6/n) GSPA facilitates a measure called gene localization, via comparison to a uniform distribution. For PBMCs, cell embeddings using all genes versus only top localized genes showed similar structure, suggesting that localized genes capture cell-type variation and geometry.
21.12.2024 17:52 — 👍 1 🔁 0 💬 1 📌 0
(5/n) We show that for (PBMCs), GSPA with compressed wavelets grouped cell-type specific genes from PanglaoDB. In embryoid-body lineages early trajectory genes linked to embryonic stem cells (NANOG, POU5F1) and late trajectory genes linked to hemangioblast-specific signatures (CD34, PECAM1, TAL1).
21.12.2024 17:50 — 👍 1 🔁 0 💬 1 📌 0
(4/n) Rather than transposing the cellxgene matrix for gene embeddings, we frame genes as signals on a cell–cell graph, decomposed using graph-based diffusion wavelets. Diffusion wavelets use differences of powers of P. Then we then reduce dimensionality with an autoencoder.
21.12.2024 17:47 — 👍 2 🔁 0 💬 1 📌 0(3/n) Low dimensional embeddings like UMAP, PHATE, tSNE have been vital in understanding geometry and subpopulations from single cell data. Gene expression is also highly organized (pathways etc). But due to noise, directly apply these embeddings to genes has not worked.
21.12.2024 17:47 — 👍 1 🔁 0 💬 1 📌 0(2/n)This work is lead by Aarthi Venkat, Sam Leone. We couldn't have done it without our collaborators Scott Youlton, Eric Fagerberg, John Attanasio from Nik Joshi Lab at Yale and Michael Perlmutter from Boise State University!
21.12.2024 17:47 — 👍 1 🔁 0 💬 1 📌 0