Home - Trost Lab
I have a 3-year postdoc position available at University of Manchester for a mass spec expert in proteomics. The person will be involved in exciting projects in single cell proteomics, drug discovery and innate immunity - on timsTOF Ultra AIP & HF & Astral Zoom.
www.jobs.ac.uk/job/DQJ093/r...
02.02.2026 19:52 — 👍 24 🔁 27 💬 0 📌 0
Rumours of 'more news in spring', which likely means no rounds until summer and no money being released until 2027 at the earliest. Given the virtual cancellation of the current round, where applicants and reviewers have already poured thousands of hours of work and effort, that's a 12 month gap.
01.02.2026 10:57 — 👍 3 🔁 3 💬 1 📌 0
VerteBrain reveals novel neural and non-neural protein assemblies conserved across vertebrate evolution
Protein-protein interactions underlie core brain functions, including neurotransmitter release, receptor activation, and intracellular signaling essential for learning, memory, and cognition. Here, we systematically map conserved brain protein interactions across five vertebrate species-rabbit, chicken, dolphin, pig, and mouse-using co-fractionation and immunoprecipitation mass spectrometry. From 2,197 biochemical fractions, we identify over 82,000 high-confidence interactions among 6,108 conserved proteins. This interaction map (VerteBrain) reveals both regulatory and structural complexes, including extensive synaptonemal protein associations likely involved in inter-neuronal coordination. Conservation across species underscores essential roles in neuronal and glial function, as well as in additional tissues for more widely expressed complexes. The VerteBrain dataset also uncovers candidate disease mechanisms, including roles for ARHGEF1 in short stature syndromes, synaptic vesicle trafficking complexes in epilepsy, and RELCH in congenital deafness. VerteBrain provides a publicly accessible framework for investigating brain protein interactions and their relevance to human neurological disorders.
(BioRxiv All) VerteBrain reveals novel neural and non-neural protein assemblies conserved across vertebrate evolution: Protein-protein interactions underlie core brain functions, including neurotransmitter release, receptor activation, and intracellular signaling essential for… #BioRxiv #MassSpecRSS
28.05.2025 21:04 — 👍 1 🔁 1 💬 0 📌 0
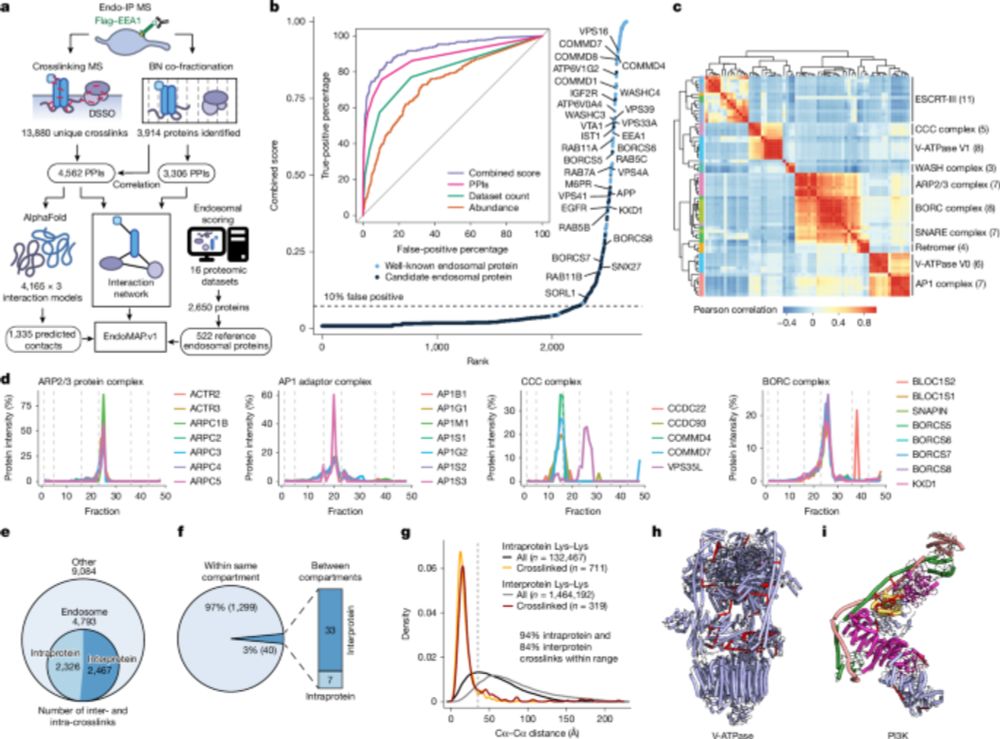
EndoMAP.v1 charts the structural landscape of human early endosome complexes - Nature
A study presents EndoMAP.v1, a resource that combines information on protein interactions and crosslink-supported structural predictions to map the interaction landscape of early endosomes.
New work @harvard by Miguel Gonzalez-Lozano @harperlabhms.bsky.social & @ernstschmid.bsky.social in Johannes Walter lab charts structural interactome of endosomes. #XL-MS #Alphafold Funded by @asapresearch.parkinsonsroadmap.org & NIH. Science continues-despite attacks www.nature.com/articles/s41...
28.05.2025 15:52 — 👍 44 🔁 19 💬 0 📌 1
