
One for the scientists
My site for calculation of recipes for thermodynamically correct pH buffers has data about the buffers that are made.
phbuffers.org/BuffferCalc/...
Analysis of about 40,000 buffers. -> lot of the five most common buffer species. Anyone care to guess what these top 5 are?
15.08.2025 20:49 — 👍 1 🔁 0 💬 1 📌 0
As a welshman, I appreciate the Dylan Thomas, albeit mutated :)
11.07.2025 18:38 — 👍 1 🔁 0 💬 1 📌 0
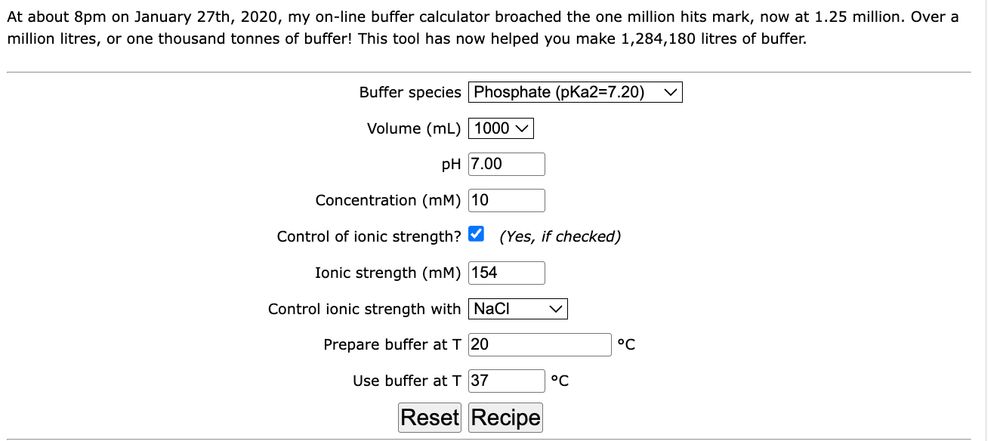
Many of you (say, about one million) have used my buffer site to calculate thermodynamically correct buffers. Well, today, I upgraded the site with added security, becuase it was stopping soem people connecting. Should all be fine now. https rather than http. Enjoy!
phbuffers.org/BuffferCalc/...
11.07.2025 18:38 — 👍 6 🔁 1 💬 1 📌 0

Just before the Beynfest! Claude continues to impress. I asked it to write a simple protein dotplot tool to explain to a PhD student how two sequences are related, and 20 minutes later:
phbuffers.org/Claude/DotPl...
Free for all to use...
07.07.2025 12:19 — 👍 2 🔁 0 💬 0 📌 0
Slight ambarassed to be at the ’Putting Beynon out to pasture’ celebration at @ukbspr.bsky.social tomorrow. Thanks to the organisers…
06.07.2025 21:31 — 👍 10 🔁 3 💬 1 📌 1
PeptideMapper Annotation Syntax Guide
I asked Claude to writ ethe annotation guide, and it built in features I did not ask for, and did not know about! I hope this will actually be of use..
phbuffers.org/Claude/pepti...
25.06.2025 21:40 — 👍 0 🔁 0 💬 0 📌 0
yes, me too. frequent saves, and 'start over' with that version. The dialogue with Claude, from the tool, actually talked about protein chemistry! I, and I suspect you, and the future generation will learn to give clear instructions.
25.06.2025 21:40 — 👍 0 🔁 0 💬 1 📌 0
PeptideMapper Annotation Syntax Guide
Here's the annotation language for peptidemapper
phbuffers.org/Claude/pepti...
25.06.2025 21:36 — 👍 1 🔁 0 💬 0 📌 0
PeptideMapper by Rob Beynon
BTW, 1600 lines of code. SO impressed
phbuffers.org/Claude/pepti...
25.06.2025 21:31 — 👍 0 🔁 0 💬 0 📌 0


"I Claudiused" - tragic tale.
My tool is now silly. (haven't quite made a mess-alina of it though!). Has a flexible annotation language, which was not easy to get right. But "P14 sequence blue out above L1" means "Label P14 with its sequence in blue outside the map" @catfranco.bsky.social
25.06.2025 21:27 — 👍 1 🔁 0 💬 2 📌 0
PeptideMapper by Rob Beynon
Oh yes, the URL of the peptide mapper:
phbuffers.org/Claude/pepti...
24.06.2025 16:52 — 👍 2 🔁 1 💬 0 📌 0

So, I rewrote my little peptide mapping tool
And, in conversation with @catfranco.bsky.social she suggested a feature - so, in 5 minutes, it was implemented.with an annotation language
K fill cyan
C circle yellow in
V diamond green out below
P45 fill orange
Fun!
24.06.2025 09:13 — 👍 2 🔁 0 💬 0 📌 0
PeptideMapper by Rob Beynon
And here it is. OK, there is no doubt that writing the original taught me how to specify the task, and there is less functionality in some areas, but more in others, yet, within about 6h, here it is! phbuffers.org/Claude/pepti...
24.06.2025 07:43 — 👍 1 🔁 0 💬 0 📌 0
I wrote a neat little tool to draw peptide maps a long time (In Livecode/Hypercard) and in 4h have had Claude create a near identical web app...when it works well I'll share a hyperlink. @catfranco.bsky.social
23.06.2025 19:04 — 👍 3 🔁 0 💬 2 📌 0
I asked it to ‘prettify’ my buffer program.. made a total mess of it over and over again…so not perfect
21.06.2025 07:13 — 👍 1 🔁 0 💬 1 📌 0
Neat tool though… and it can always be improved . In fact hard to leave alone..
21.06.2025 07:11 — 👍 0 🔁 0 💬 1 📌 0
Loved it.. now you know where ‘MEERCAT’ came from…
21.06.2025 07:09 — 👍 1 🔁 0 💬 0 📌 0
Loved it! We are still looking for marilin- watch this space! Fantastic interview! Will see Kathryn in the BSPR meeting in Liverpool in July, www.bspr.org/annual-meeti... when there is a ‘putting Beynon out to pasture’ session! Payback for the embarassment!
21.06.2025 07:07 — 👍 2 🔁 0 💬 0 📌 0
neat - screenshot?
19.06.2025 20:37 — 👍 0 🔁 0 💬 1 📌 0
"vibe coding" 🤮 😂
19.06.2025 13:55 — 👍 0 🔁 0 💬 0 📌 0
capsaicin? Cathepsin is a cysteine proteinase e.g. papain, bromelain, so maybe in papaya and pineapple? :)
19.06.2025 08:50 — 👍 1 🔁 0 💬 1 📌 0
.
19.06.2025 08:49 — 👍 0 🔁 0 💬 0 📌 0
Any other stories like this? end/ Anyone used Ai to code scientific applications? I do think the main limitation is in specifying the parameters of the tools that I want to make. That is on me. end/
19.06.2025 08:49 — 👍 3 🔁 0 💬 4 📌 0
But, for ordinary, inept folk like me, this makes simple tasks completely within reach. e.g. I asked Claude for an SVG viewer for my peptide mapper programme -easy. Of course, I will now rewrite the entire tool as a web app.. so much more user friendly. 8/
(phbuffers.org/Research/Pma...)
19.06.2025 08:49 — 👍 0 🔁 0 💬 1 📌 0
Scarily, Claude not only correctly interpreted the Python code but also intimated that if I had the exe file, it could probably reverse engineer that too. 7/
19.06.2025 08:49 — 👍 0 🔁 0 💬 1 📌 0
And bingo, a web app that replaces an exe file with a fast and pretty, user-friendly flexible web app. I did no coding at all, and it took me less than a day of intermittent access. The decoding is instantaneous for large files. 6/
19.06.2025 08:49 — 👍 0 🔁 0 💬 1 📌 0
Breakthrough. A very clever research in our group had previously decoded the data, and we found the old Python code to do that. Showed this to Claude and bingo! Immediate result. After that, long dialogue about making it pretty, export options, tooltips and an explanation of the coding method 5/
19.06.2025 08:49 — 👍 0 🔁 0 💬 1 📌 0
So, Claude tried to build a lookup table to reverse engineer the time from known examples. Many days of data, at very high time resolution, was never going to happen, as it required just too many data points, and even then, no guarantee all possibilities would be covered. 4/
19.06.2025 08:49 — 👍 0 🔁 0 💬 1 📌 0
But we hit an interesting sticking point. Claude correctly identified which bytes encoded the timebase, but could not unpack the highly compressed format (it involved a variable number of sixteenths of a second increments) 3/
19.06.2025 08:49 — 👍 0 🔁 0 💬 1 📌 0
The first attempt was poor, and with a bit of inspection, I was able to let Claude know what the EOF and EOR markers were, as well as indication what the columns of data were when unpacked. Second attempt - so much better. 2/
19.06.2025 08:49 — 👍 0 🔁 0 💬 1 📌 0
Ras biologist at the University of Liverpool when I’m not trying to lead Faculty provision of core facilities and infrastructure improvements.
Green Party activist with Trafford Green Party
Co-greyhound pawrent 🐶 with @GreenPartyHan
Em. Prof of Molec Microbiology & former Vice-Principal, Univ Edinburgh. FLSW, FRSE, etc.. Allegedly retired, but still doing science-related stuff. Trustee FairFrome charity. Pro-EU Brit. #iPhobe. Editor #AccessMicrobiology
Image analyst at the Centre for Cell Imaging, the University of Liverpool.
didyouknowimageanalysis.wordpress.com
CCI.liv.ac.uk
Precision-function-based microbiome trials.
Ex vivo microbiome assays.
Microbiome–xenobiotic interactions.
Meta-omics analyses
Bayerisches Zentrum für Biomolekulare Massenspektrometrie | TUM School of Life Sciences | Proteomics | Metabolomics | Bioinformatics | 🇩🇪
The official channel for the British Society for Proteome Research
MRC PhD student at University of Manchester
Targeted proteomics for biomarker discovery/validation in pancreatic cancer
Virologist, bioinformatician, NGS, photographer.
RNA is my special interest 🧬
Proteomics is growing on me 😬
🇳🇿🇬🇧British Kiwi based in Liverpool 📍
Post doc in the Emmott Lab👩🏻🔬👩🏻💻
🏳️🌈 She/They.
🧠 Neurodivergent ✨
rebee.co.uk
Mass Spectrometry and proteomics at Uni of Manchester. Analysis of primary samples for biomarker discovery. Development of targeted assays for clinical use.
Co-founder, Complement Therapeutics
Also football (SWFC), cricket and politics.
Proteomics lab in Sheffield (UK) studying protein interactions and PTMs in health and disease with a particular interest in neuroproteomics.
Ex fun-guy; now a research professional with Muppet tendencies. Views are my own...
Zoologist turned psychologist. I study behaviour, smell, and maternal-infant interaction at the Universities of Stirling and Wroclaw. Učím se česky. 🇺🇦 🇪🇺 🏴 🇨🇿 🇵🇱
www.scraigroberts.com
Tenure Track Fellow, Kidney Research UK Senior Non-Clinical Fellow. Analytical Chemistry, biological/inorganic mass spectrometry. Proteomics, glycans, metabolomics.
Systems biologist • Prof at Max Perutz Labs & MedUni Vienna • dad×2 • he/him
#LoveVirology #TeamMassSpec #dataviz
📍 Vienna 🇪🇺
🇲🇽🇫🇷🇪🇺 Analytical chemist, specialized in #MassSpec #Proteomics. Leading the Immunopeptidomics Platform at HI-TRON Mainz / DKFZ
PhD in Biochemistry and Biotechnology
University of Glasgow
PostDoc in Olsen Group and Arachnid wll-being executive manager. 🕷️🕸️ Working on Spatial Proteomics and extracellular vesicles. 🤗





